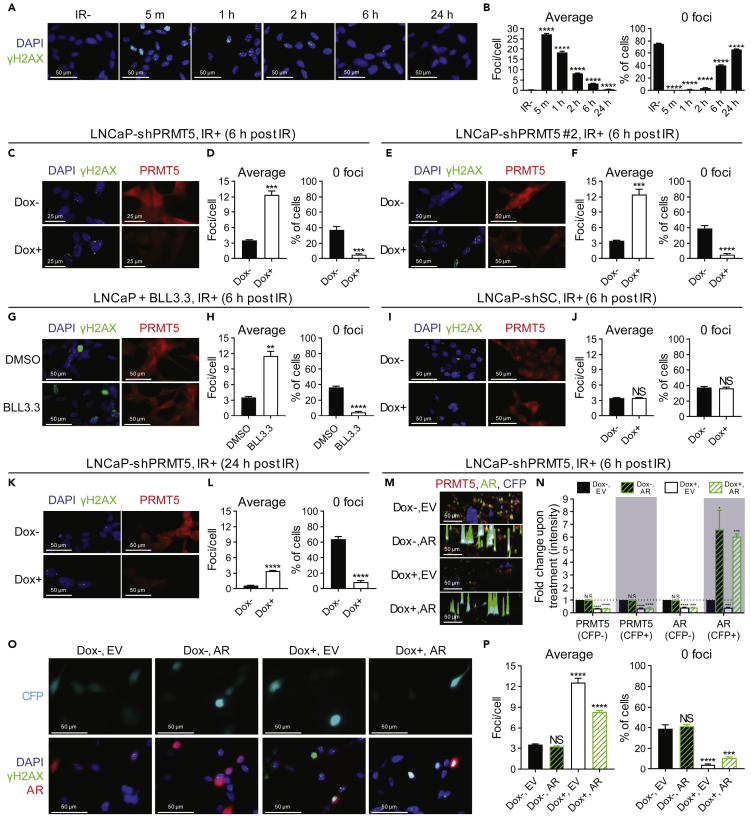
Figure 2.
PRMT5 Regulates the Repair of DNA Double-Strand Breaks in Prostate Cancer Cells Independently of AR Expression
(A) Time course of the formation and repair of DSBs (γH2AX foci) at the indicated minutes (m) or hours (h) post 2 Gy IR in LNCaP cells.
(B) Quantification of DSBs in each individual cell from A: “average” indicates the average number of DSBs in each cell and “0 foci” indicates the percentage of cells that do not contain any DSBs.
(C, E, G, I, and K) DSBs 6 h or 24 h post 2 Gy IR in the indicated cells (C: LNCaP-shPRMT5, E: LNCaP-shPRMT5 #2, G: LNCaP, I: LNCaP-shSC, K: LNCaP-shPRMT5) with (Dox+) and without (Dox−) PRMT5 knockdown/scramble control (SC) knockdown or with (BLL3.3) and without (DMSO) PRMT5 inhibition.
(D, F, H, J, and L) Quantification of DSBs from C, E, G, I, and K as described above in Figure 2B (C: LNCaP-shPRMT5, E: LNCaP-shPRMT5 #2, G: LNCaP, I: LNCaP-shSC, K: LNCaP-shPRMT5).
(M) LNCaP-shPRMT5 cells were co-transfected with plasmids encoding Flag-AR (AR) or empty vector (EV) and a plasmid encoding cerulean fluorescent protein (CFP). Fluorescence images acquired 6 h post 2 Gy IR are representative immunocytochemistry images in 3D where each peak is a cell and the height of each peak is the intensity of signal. Blue peaks represent transfected CFP-expressing cells (CFP+). Colors are indicated as follows: endogenous PRMT5 (red), endogenous and exogenous AR (green), and exogenous CFP (cerulean).
(N) Quantification of protein intensity from M in untransfected (CFP−) and transfected (CFP+) cells. For each biological replicate, values were normalized to the value for “Dox−/EV” to calculate the fold change in protein expression upon treatment.
(O) DSBs at 6 h post 2 Gy IR in LNCaP-shPRMT5 cells where AR expression was rescued via co-transfection with plasmids encoding Flag-AR (AR) or empty vector (EV) and a plasmid encoding Cerulean fluorescent protein (CFP) as a transfection control.
(P) Quantification of DSBs in transfected cells (defined as CFP+) from O as described above.
Fluorescence images in A, C, E, G, I, and K are representative immunocytochemistry images (blue = DAPI, green = γH2AX, red = PRMT5). Fluorescence images in M are representative immunocytochemistry images (red = PRMT5, green = AR, and blue = CFP). Fluorescence images in O are representative immunocytochemistry images (blue = DAPI, green = γH2AX, red = AR, Cerulean = CFP). All bars are the mean ± s.d. of four independent experiments. Statistical analysis for B, N, and P comparing experimental with the control (“IR−”, “Dox−”, “DMSO”, or “Dox−/EV”) was performed using Brown-Forsythe and Welch ANOVA followed by Dunnett's T3 multiple comparisons test, whereas statistical analysis for D, F, H, J, and L comparing experimental with the control (“Dox−” or “DMSO”) was performed using Welch's t test (*p ≤ 0.05; **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, NS p > 0.05).