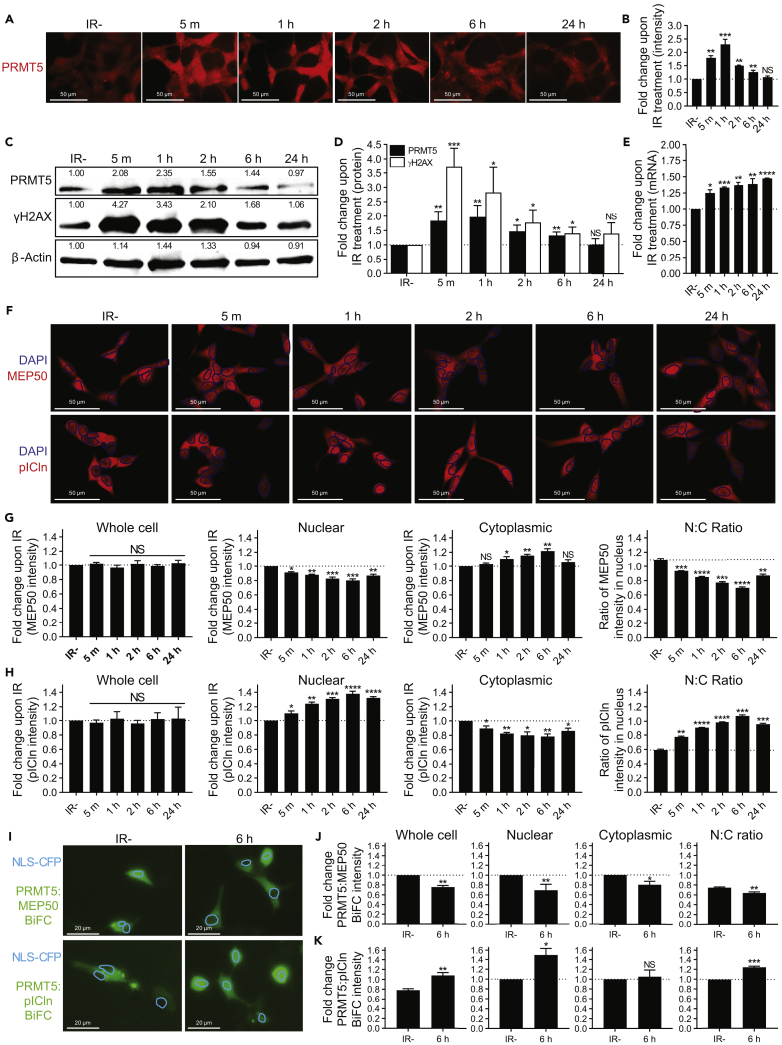
Figure 6.
IR Induces PRMT5 Expression, pICln Nuclear Localization, and the PRMT5:pICln Interaction in the Nucleus
(A) Time course of PRMT5 expression at the indicated minutes (m) or hours (h) post 2 Gy IR in LNCaP cells.
(B) Quantification of PRMT5 expression in images from A. For each biological replicate, values were normalized to the value for “IR−” to calculate the fold change in protein expression upon IR.
(C) Representative Western blot showing the time course of protein expression at the indicated minutes (m) or hours (h) post 2 Gy IR in LNCaP cells. Values shown indicate the intensity relative to IR− for the biological replicate used as the representative Western blot.
(D) Quantification of protein expression via Western blotting from C. For each biological replicate, values were normalized to the value for “IR−” to calculate the fold change in protein expression upon IR.
(E) Time course of PRMT5 expression at the mRNA level at the indicated minutes (m) or hours (h) post 2 Gy IR in LNCaP cells via RT-qPCR. For each biological replicate, values were normalized to the value for “IR−” to calculate the fold change in mRNA expression upon IR.
(F) Time course of MEP50/pICln expression/localization at the indicated minutes (m) or hours (h) post 2 Gy IR in LNCaP cells.
(G) Quantification of MEP50 expression/localization in images from F: “Whole cell” indicates MEP50 expression in the entire cell, “Nuclear” indicates MEP50 expression in the nucleus, which was defined by DAPI staining, “Cytoplasmic” indicates MEP50 expression in the cytoplasm that was defined as staining outside DAPI, and “N:C ratio” was calculated by dividing the value for nucleus by the value for cytoplasmic for each cell individually such that an N:C ratio of 1 indicates equal expression in both the nucleus and cytoplasm.
(H) Quantification of pICln expression/localization in images from F as described above.
(I) PRMT5:MEP50 and PRMT5:pICln interaction 6 h post 2 Gy IR in irradiated (6 h) and non-irradiated (IR−) LNCaP cells via BiFC assay.
(J) Quantification of PRMT5:MEP50 BiFC intensity in images from I: “Whole cell” indicates BiFC intensity in the entire cell, “Nuclear” indicates BiFC intensity in the nucleus that was defined by NLS-CFP signal, “Cytoplasmic” indicates BiFC intensity in the cytoplasm that was defined as staining outside NLS-CFP signal, and “N:C ratio” was calculated by dividing the value for nucleus by the value for cytoplasmic for each cell individually such that an N:C ratio of 1 indicates equal interaction in both the nucleus and cytoplasm. NLS-CFP was used as a transfection control and a marker of the nucleus.
(K) Quantification of PRMT5:pICln BiFC intensity in images from I as described above.
Fluorescence images in A are representative immunocytochemistry images (red = PRMT5). Fluorescence images in F are representative immunocytochemistry images (blue = DAPI and red = MEP50 or pICln). Blue circles outline DAPI staining to allow for better visibility of expression in the nucleus. Fluorescence images in I are representative images from BiFC assay (green = PRMT5:MEP50 and PRMT5:pICln, cerulean = NLS-CFP). Blue circles outline NLS-CFP signal to allow for better visibility of expression in the nucleus. All bars are the mean ± s.d. of three independent experiments. Statistical analysis for B, D, E, G, and H comparing experimental with the control (“IR−”) was performed using Brown-Forsythe and Welch ANOVA followed by Dunnett's T3 multiple comparisons test, whereas statistical analysis for J and K comparing experimental with the control (“IR−”) was performed using Welch's t test (*p ≤ 0.05; **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, NS p > 0.05).