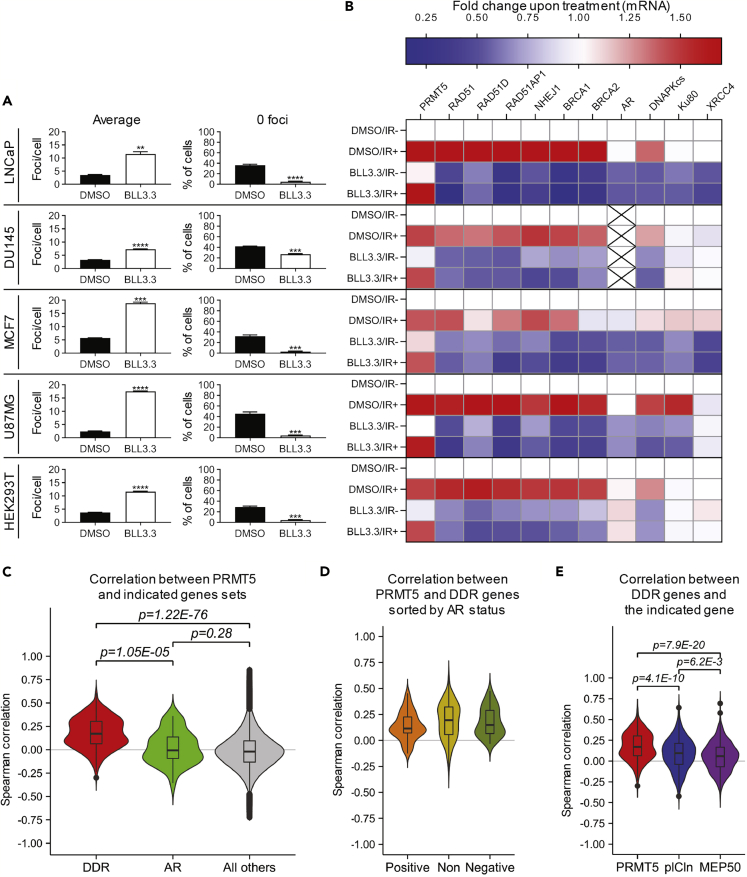
Figure 7.
PRMT5 Regulates the Expression of DDR Genes in Multiple Cancer Cell Lines, and PRMT5 Expression Positively Correlates with DDR Genes in Human Cancer Tissues
(A) Quantification of DSBs 6 h post 2 Gy IR in the indicated cell lines with (BLL3.3) and without (DMSO) PRMT5 inhibition as described in Figure 2B (see also Figures S7A–S7E for representative images).
(B) Quantification of mRNA via RT-qPCR 6 h post 2 Gy IR in the indicated cell lines with (BLL3.3) and without (DMSO) PRMT5 inhibition. For each biological replicate, values were normalized to the value for “DMSO/IR−” (untreated) to calculate the fold change in mRNA expression upon treatment (see also Figures S7F–S7J for statistical analysis).
(C) Violin plots representing Spearman correlations comparing the mRNA expression level between PRMT5 and DDR genes (DDR), PRMT5 and AR (AR), or PRMT5 and all other genes (All other) across 32 clinical cancer datasets from TCGA. The gene set for DDR genes was defined as RAD51, RAD51D, RAD51AP1, NHEJ1, BRCA1, BRCA2, WEE1, DNAPKcs, Ku70, Ku80, an XRCC4 (see also Figure S8).
(D) Violin plots representing Spearman correlations comparing the mRNA expression level between DDR genes and PRMT5. Cancer types were stratified by the correlation coefficient (c.c.) between PRMT5 and AR: positively correlated (c.c. > 0 & p < 0.01) (Positive), negatively correlated (c.c. < 0 & p < 0.01) (Negative), or not correlated (p > 0.01) (Non).
(E) Violin plot representation of Spearman correlation values between DDR genes and either PRMT5, CLNS1A (pICln), or WDR77 (MEP50).
Bars in A and values used in the heatmap in B are the mean of three independent experiments. Box-and-whiskers plots in C–E show the median value (line) and interquartile range between the first and third quartiles (box). The upper whisker extended to the largest value no further than “1.5 x interquartile range” and the lower whisker extended to the smallest value at most “1.5 x interquartile range”. Outliers beyond the whiskers are shown as dots. Statistical analysis in A comparing experimental to the control (“DMSO”) was performed using Welch's t test (**p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001). Statistical analysis in C–E was performed using Wilcoxon test, and the p values are displayed.