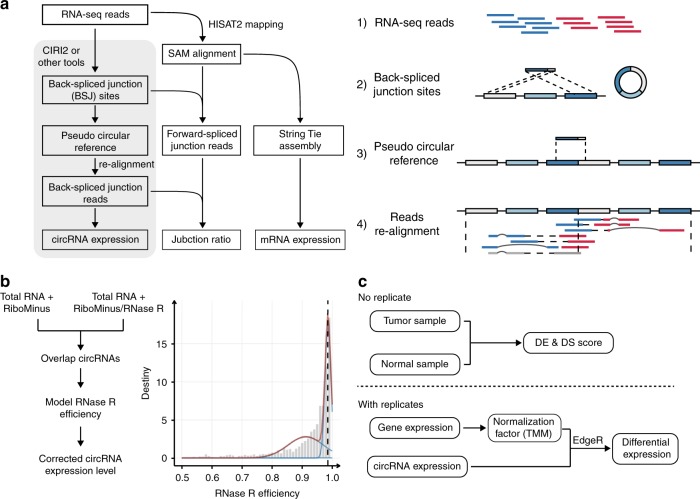
Fig. 2. Overview of the CIRIquant pipeline.
a Workflow of circRNA quantification. CircRNAs are first identified by CIRI2 or any other computational methods. Then, CIRIquant detects back-spliced junction reads using re-alignment of RNA-seq reads against the pseudo-circular reference. Expression level and junction ratio of circRNAs are calculated using BSJ reads and FSJ reads, and stringTie is used to estimate gene abundance. b The correction of RNase R treatment efficiency. Overlapped circRNAs detected in both RiboMinus and RiboMinus/RNase R data are used for enrichment efficiency calculation and a gaussian mixture model (GMM) is used for model fitting. The fitted model is used as prior distribution for the correction of circRNA expression values. c Differential expression analysis of circRNAs. Top: For paired samples without replicates, DE-score and DS-score is directly calculated using generalized fold change. Bottom: For paired samples with biological replicates, gene expression values are used to calculate normalization factor, then the modified edgeR pipeline is used for statistical significance test.