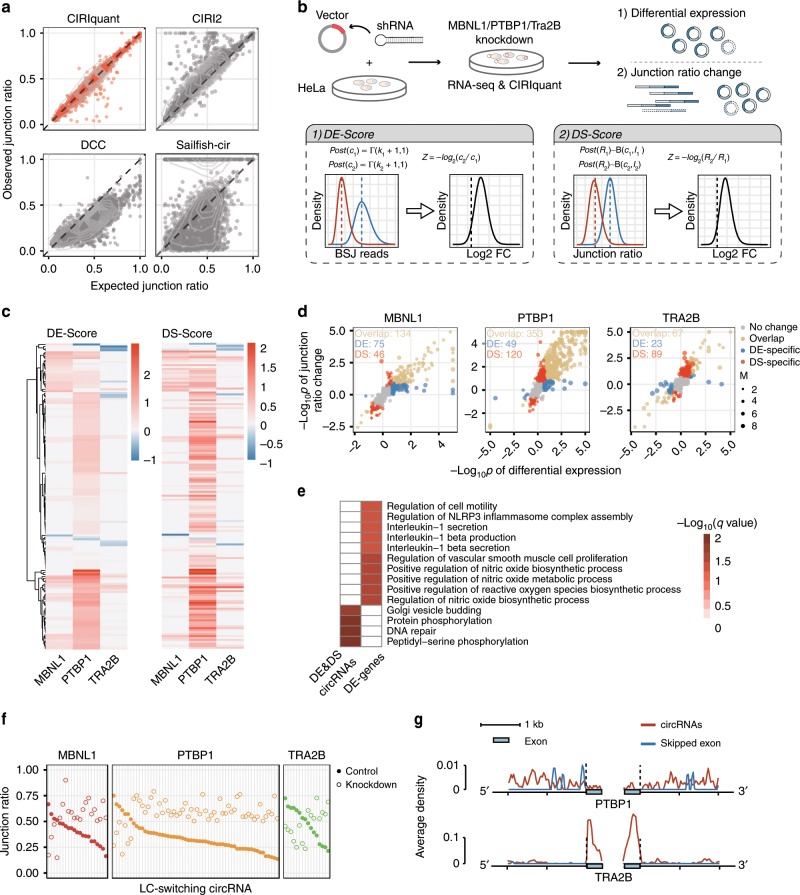
Fig. 5. Differentially spliced circRNAs after knockdown of three splicing factors.
a The correlation between simulated and estimated junction ratio by CIRIquant and three other methods. x- and y-axis represent the expected and estimated junction ratio of circRNAs, respectively. For DCC, the junction ratio was calculated using the results from LinearCount and CircRNACount output. For Sailfish-cir, the TPM of circRNAs and corresponding host genes were used for junction ratio calculation. b Schematic view of experimental design for shRNA induced knockdown of three splicing factors (MBNL1, PTBP1, and Tra2B) and the calculation of DE- and DS- score. Specially designed shRNAs were used to knockdown target genes. Then, RNA-seq were performed for control and knockdown groups, and circRNA expression levels and junction ratio were estimated by CIRIquant for further differential expression analysis. c Heatmap of DE score and DS score in three knockdown datasets. Only circRNAs significantly changed in at least one sample are shown in the heatmap. d Overlap of significantly changed circRNAs. x- and y-axis represent −log10(p value) from DE-analysis and rate-ratio test using junction ratio of circRNAs in two different conditions, respectively. Downregulated p-values are assigned as negative values. DE-specific and DS-specific circRNAs are highlighted in blue and red, respectively. e Gene ontology enrichment analysis of differentially expression genes and all significantly changed DE- and DS-circRNAs. The logarithm of FDR was calculated in Enrichr, then plotted in heatmap for comparison. f Junction ratio changes of LC-switching circRNAs after MBNL1/PTBP1/TRA2B knockdown. g Average depth of binding sites in flanking regions of differentially spliced ES exons and significantly changed circRNAs using CLIP-seq results from iCLIPdb. A sliding window analysis with 8 bp windows was performed for density calculation. Vertical axis represents the average binding site per shifted window.