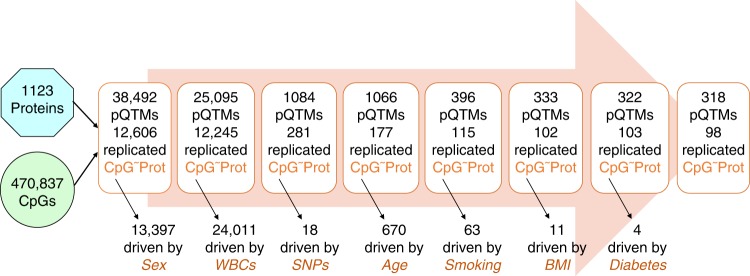
Fig. 2. Summary of the step-wise EWAS.
A series of pEWASs was carried out in the discovery cohort KORA, followed by replication in the QMDiab study (Supplementary Data 2 and 3). In the initial step, methylation levels of 470,837 CpGs (M-values, winsorized) were tested for association with 1123 blood circulating protein concentrations (log-scaled, winsorized), leading to 38,492 associations that reached stringent Bonferroni significance, 12,606 of which were replicated. Potential driving factors (sex, white blood cell composition, genetic variants, age, smoking, BMI, and diabetes) were successively regressed out from the CpG and the protein levels, using the residues in the next EWAS step (see Methods). At each step, a number of associations (pQTMs) fell below the significance threshold (indicated by the black arrows). These associations were likely driven by the factor used in the previous regression step. Eventually, 318 pQTMs remained that were not driven by any of the factors listed here, 98 of which were replicated (Table 1).