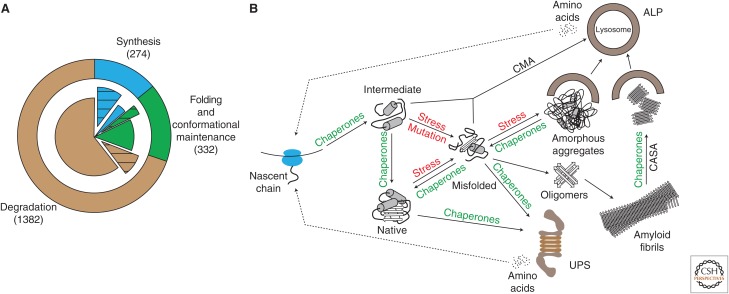
Figure 1.
Architecture of the proteostasis network (PN). (A) The three modules of the PN (outer ring): synthesis (blue, 274 factors), folding and conformational maintenance (green, 332 factors), and degradation (brown, 1382 factors). The inner pie chart shows the partitioning of each module into essential (hatched sectors, synthesis [203/274], folding [78/332], and degradation [178/1382]) and nonessential components (filled sectors). (B) Central role of molecular chaperones in the PN. The nascent polypeptide chain emerging from the ribosomal tunnel is prevented from misfolding and aggregation by chaperones, including components that bind to the ribosome (trigger factor in bacteria, nascent chain–associated complex [NAC] and ribosome-associated complex [RAC] in eukarya). A second tier of chaperones (Hsp70, Hsp90, chaperonins) does not interact with the ribosome directly and mediates co- or posttranslational folding. Proteins that misfold because of mutations or under conditions of stress are selectively degraded either by the ubiquitin–proteasome system (UPS) or chaperone-mediated autophagy (CMA) and chaperone-assisted selective autophagy (CASA). Misfolded proteins may aggregate to soluble oligomers, amorphous aggregates, or amyloid fibrils when basal chaperone and degradation capacity is exceeded. Aggregates may be sequestered into insoluble inclusions, which may be dissociated into fragments by specialized chaperones (Hsp104 in yeast, Hsp70/Hsp40/Hsp110 in metazoans) for subsequent clearance by the autophagy–lysosomal pathway (ALP).