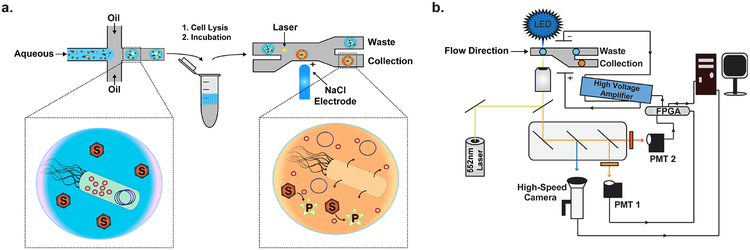
Figure 1. Fluorescence-activated droplet sorting.
(a) Overview of microfluidic droplet sorting. A library of genes encoding a specific protein or enzyme are expressed in E. coli and encapsulated in microfluidically generated w/o droplets along with a sensor and corresponding substrates. Expressed proteins are liberated from their E. coli host via chemical, enzymatic, or heat lysis and incubated at the desired reaction temperature off-chip. Following incubation, the droplet population is sorted using the FADS system to enrich for members with a desired functional activity. (b) Overview of the optical train and electronic components of the FADS system. For Cy3 based fluorescence, incident light from a 552 nm laser is focused onto the sorting junction of the microfluidic chip through a 20x objective to excite active droplets. Emitted light is led back through the objective past a series of dichroic mirrors to a PMT with a 563 nm band-pass filter. The signal from the PMT is fed to a FPGA controlled by custom LabView software. Upon detecting a fluorescent signal, the FPGA is instructed to energize a 4M NaCl electrode. A high voltage amplifier amplifies the generated sorting pulse to produce the DEP force necessary to deflect the target drop into the collection channel. Blue light with minimal Cy3 spectral overlap is used to illuminate the sorting chip for monitoring with a high-speed camera. A host PC interfaces with the FPGA to collect data and adjust sorting parameters.