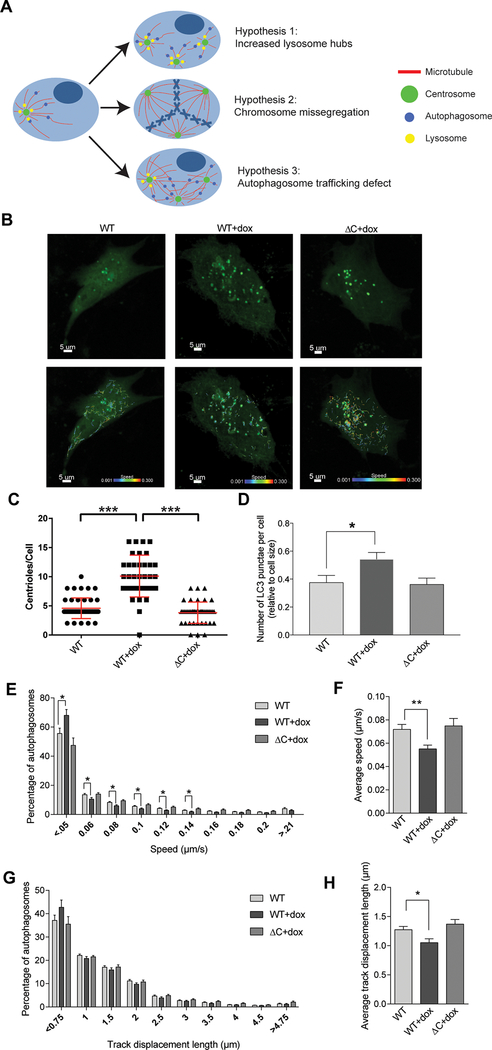
Figure 4: Centrosome amplification impairs autophagosome trafficking.
(A) Diagrammatic representation of three potential hypotheses for disrupted autophagy in cells with centrosome amplification. In the first scenario, centrosome amplification results to a disordered array of microtubules, thereby decreasing the efficiency of transportation of autophagic vesicles in the cell. In the second scenario, an increase in centrosomes generates an increase in lysosomes hubs, as lysosomes tend to cluster around centrosomes. In the third scenario, centrosome amplification causes chromosome missegregation, resulting in aneuploidy and therefore altered stoichiometry of protein complexes, increasing the cell’s dependence on autophagy. (B) Cells were plated in glass-bottom chamber slides, treated with doxycycline (if indicated), and transduced with baculovirus encoding LC3-GFP to label autophagosomes. Timelapse imaging was used to measure autophagosome trafficking. Representative images of RPE cells are shown both with and without the autophagosome traces. Scale bar = 5 μm. Colored lines on the bottom images correlate with autophagosome speed (color of line) and distance (length of line). (C) Following imaging, cells were fixed and stained to assess CA, which is quantified in this panel by counting centrioles (centrin foci). Dots represent individual cells. (D) Quantification of the number of autophagosomes (LC3B punctae) per cell. (E) Histogram demonstrating the spread of autophagosome trafficking speed by condition. (F) Average autophagosome speed for each condition. (G) Histogram demonstrating the spread of autophagosome track displacement lengths by condition. (H) Average autophagosome track displacement length for each condition. Bars represent means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. NS = not significant. ANOVA and t test with Tukey’s correction for multiple comparisons were utilized. RPE cells were utilized for all experiments in this figure.