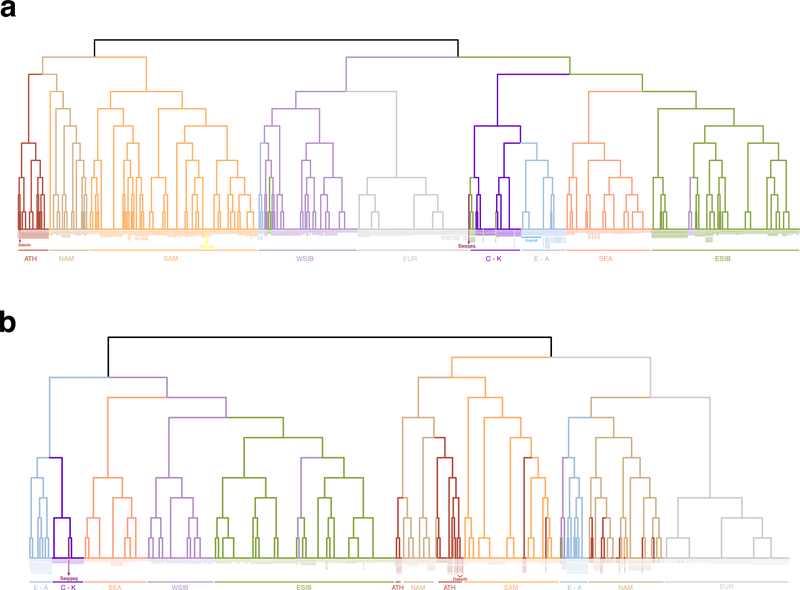
Extended Data Figure 9. Clustering trees of individuals computed by fineSTRUCTURE.
The trees are based on coancestry matrices of counts of shared haplotypes. Reduced versions of the HumanOrigins (a) and Illumina (b) SNP array datasets were used (Supplementary Table 5), including only the following meta-populations most relevant for our study: Eskimo-Aleut speakers (E-A), Chukotko-Kamchatkan speakers (C-K), Na-Dene speakers (ATH), northern First Americans or First Peoples (NAM), Southern First Peoples (SAM), West Siberians (WSIB), East Siberians (ESIB), Southeast Asians (SEA), Europeans (EUR). Meta-population affiliation is color-coded for individuals. Iñupiat individuals genotyped in this study are marked with a blue line. The two Dakelh (Northern Athabaskan) individuals with sequenced genomes and the ancient individuals, Clovis within the Southern First Peoples clade and Saqqaq within the Chukotko-Kamchatkan clade, are also indicated. Most members of each clade belong to the meta-populations indicated, with a few exceptions. First (see panel a), Altaians fall into the ESIB clade, some Chilote fall into the NAM, and Aleuts fall into the WSIB clades (two latter cases might be explained by extensive European ancestry in Chilote and in Aleuts (Extended Data Fig. 8a) which drives this clustering). Second (see panel b), some Selkups fall into the ESIB clade, all four Southern Athabaskan speakers cluster with South Americans, reflecting their substantial South American ancestry (Extended Data Fig. 8b), one Haida individual clusters with Na-Dene speakers, and five Northern Athabaskan speakers cluster with other Northern First Peoples.