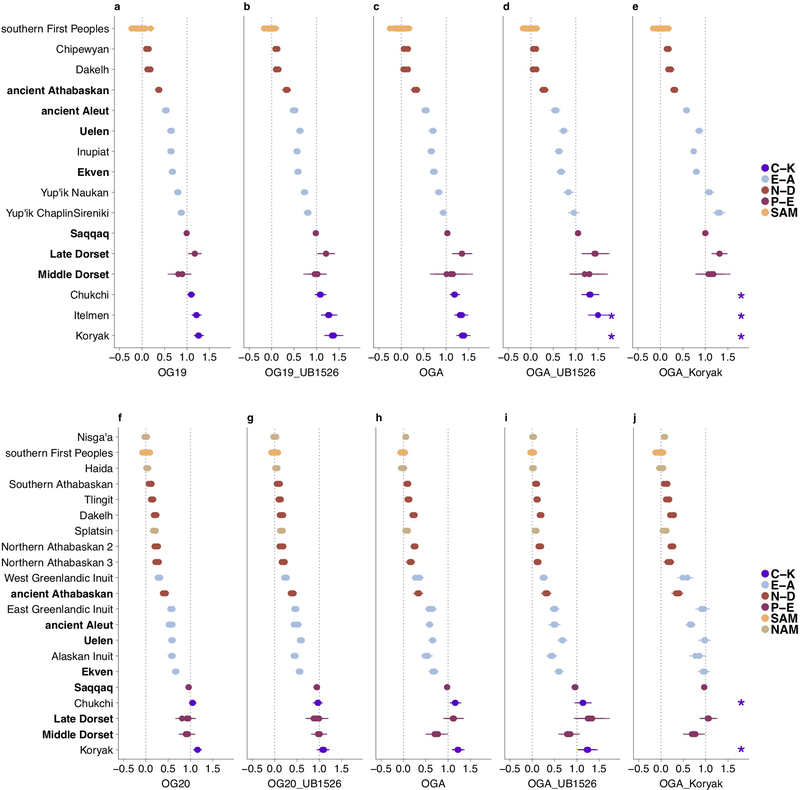
Extended Data Figure 3. Ancestry proportions in American, Chukotkan and Kamchatkan populations.
Shown are the HumanOrigins (a-e) and Illumina (f-j) datasets without transition polymorphisms. Five alternative outgroup sets are indicated below the plots and described in detail in Methods and in Supplementary Information section 5. Target populations in bold denote ancient populations. Saqqaq (pseudo-haploid genotype calls) was considered as a Paleo-Eskimo source for all populations apart from Saqqaq itself, for which Late Dorset was used as a source, and alternative First American sources were as follows: Mixe, Guarani, or Karitiana for the HumanOrigins dataset; Nisga’a, Mixtec, Pima, or Karitiana for the Illumina dataset. To visualize both systematic and statistical errors, ancestry proportions inferred by qpAdm and their standard errors are shown for all triplets including these different First Peoples sources, or for many alternative target populations in the case of Southern First Peoples (single standard error intervals are plotted here). Asterisks stand for ancestry proportions >150% (inappropriate models). Meta-populations are color-coded and abbreviated as follows: C-K, Chukotko-Kamchatkan speakers; E-A, Eskimo-Aleut speakers and ancient Neo-Eskimos and ancient Aleuts; N-D, Na-Dene speakers; NAM, Northern First Peoples; SAM, Southern First Peoples. Target population sizes in the HumanOrigins dataset ranged from 1 to 23 individuals, 5.6 on average, and in the Illumina dataset they ranged from 1 to 16 individuals, 5.1 on average.