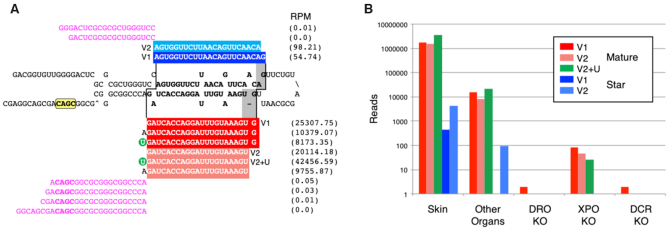
Figure 2.
The annotation of microRNA sequences and the implementation of transcriptional and processing information for each miRNA gene in MirGeneDB. (A) The structure and read stacks for Hsa-Mir-203. The precursor sequence is shown in bold; mature reads are shown in red and star reads in blue with the reads per million for each major transcript detected shown to the far right. A ‘CNNC’ processing motif (68) is shown in yellow. Also shown are the 5′ and 3′ miRNA offset reads (magenta), which clearly conform to the indicated Drosha cut (staggered line, left) given the reads processed from this locus. The Dicer cut (staggered line, right) results in two primary mature forms (dark vs light red), what we term ‘variants’ (v) that are offset from one another by 1 nucleotide (gray). The 5′ end of variant one starts with the ‘G’ whereas the 5′ end of variant two is moved 1 nucleotide 3′ and starts with the ‘U.’ Each of these two major Dicer products is accompanied by the appropriate star sequence, with variant 1 shown in dark blue and variant 2 in light blue. The mature form of variant 2—but not version 1—is heavily mono-uridylated at its 3′ end (green circle) and is thus a ‘Group 2’ miRNA (59,66). (B) The quantification of Hsa-Mir-203 read across various human-specific data sets. As expected (e.g. (72)) expression in skin is about ∼2 orders of magnitude higher relative to other organs sampled (e.g., brain, liver, stomach, lung, uterus, pancreas, testes, colorectum, small intestine and kidney) and the detection of the mature form is nearly 3 orders of magnitude relative to the star. Consistent with Mir-203 being a bona fide miRNA, expression is nearly abrogated in DROSHA and DICER knock-outs, and greatly diminished in the EXPORTIN-5 knock-out (59).