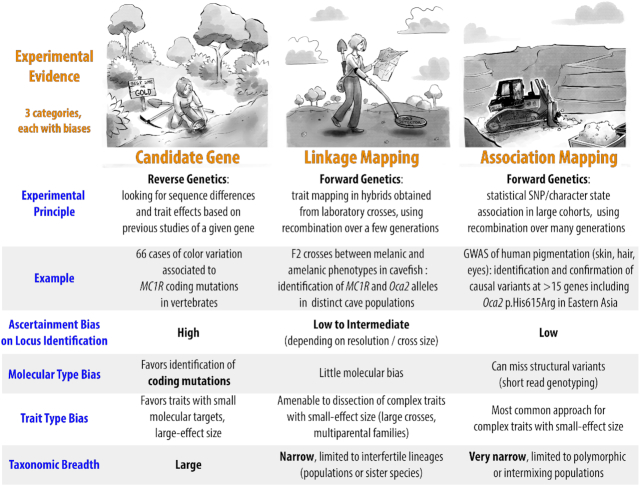
Figure 2.
Three kinds of experimental strategies for identifying gene-to-phenotype variations. Gephebase focuses on genes that have been mapped using a forward genetics approach, and supported as the causal agents by sufficient evidence. Candidate gene approaches are also included and cover broader phylogenetic distances (e.g. human/chimp), but tend to be biased towards the identification of coding changes for relatively simple traits. The search for the genes and mutations that drive phenotypic variation is somewhat analogous to searching gold: from left to right, targeted candidate gene approaches can identify variants of large effects at loci previously identified in other organisms; in a linkage mapping approach, the experimenter walks on chromosomes to narrow down the causal genetic interval, and can increase resolution and sensitivity with the analysis of more recombinants; association mapping (e.g. GWAS) takes advantage of statistical power across large datasets to extract genetic variants in linkage disequilibrium with the causal mutations. GWAS: Genome-Wide Association Studies, SNP: Single Nucleotide Polymorphism.