Figure 1.
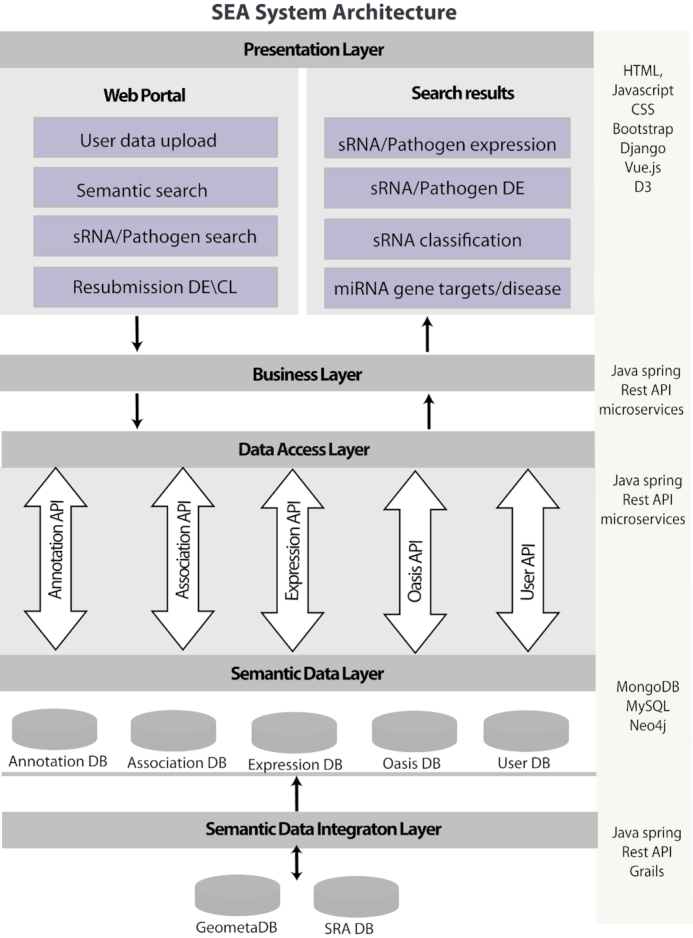
SEAweb system architecture. SEAweb system was developed using the modular system design approach (model-view-controller). The system has a presentation layer for user interface and visualization of search results. Presentation layer is followed by a business layer which transform complex user queries and distribute particular requests to the data access layer REST API services. There is a semantic data layer, to store and access primary and derived data together with annotations and links to secondary data. Annotation-DB stores metadata for experiments, samples, corresponding ontological terms as well as relations between dataset/sample, sample/term and term/term. Association-DB contains information about sRNAs and genes chromosomal locations, miRNA target genes and miRNA disease associations. Expression-DB stores sRNA expression profiles, sRNA differential expression; sRNA based classification as well as pathogen detection and pathogen differential expression. It also store details about dataset processing pipeline and parameters. Oasis-DB was used to store novel predicted miRNA information. User DB contains in-house data uploaded by the end users from Oasis 2 pipeline. Semantic data integration layer integrates primary and secondary data into the mentioned databases. Microservices were implemented in order to achieve strong encapsulation and well-defined interfaces via REST APIs.
