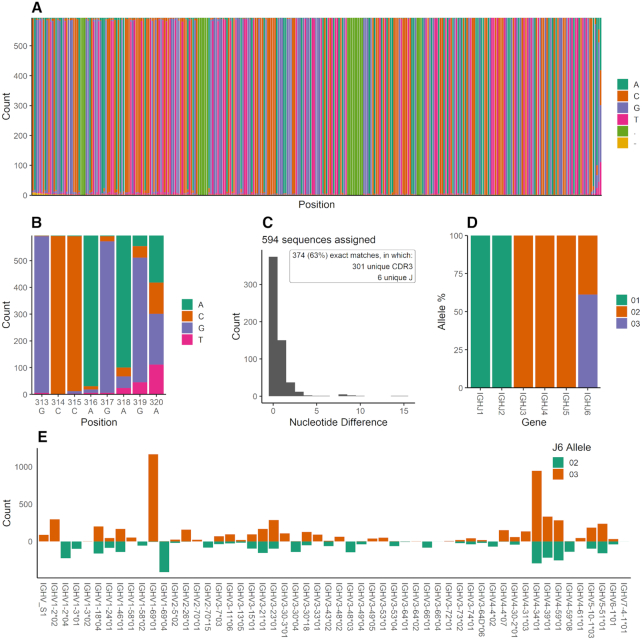
Figure 2.
Illustrative plots produced by OGRDB analysis scripts from supported inference tools. These can be provided as part of a novel allele submission, or used independently by researchers interested in exploring the usage characteristics of an AIRR-seq repertoire. (A) IMGT alignment of all sequences assigned to a novel allele (IMGT gaps shown as ‘.’, base calls with low confidence as ‘-'), showing, in this case, a high-quality underlying dataset. (B) for V-genes, a ‘zoom’ of the previous plot showing the final 8 nucleotides at the 3′ end and illustrating the effect of the recombination process on the difficulty associated with inference of the final nucleotide of the IGHV gene based on the underlying data (27) (for J-genes, a similar plot is shown of the 5′ end). (C) a histogram, similar to that produced by IgDiscover (21), showing the reads assigned to the allele, distributed by the number of nucleotide differences to the reference (or inferred reference) sequence. (D) For the analysis of novel V-genes, a plot showing the usage of J-alleles within each J-gene, which can be used to identify heterozygosity (28). In this case, the subject is heterozygous in the IGHJ6 gene. For the analysis of novel J-genes, a similar plot of V-gene allele usage is provided. (E) where heterozygosity may be present, a plot is provided showing the number of reads of each V-gene, split by their usage of the heterozygous allele (in this case alleles *02 and *03 of IGHJ6). The novel allele IGHV_S1 is found exclusively in association with the IGHJ6*03 and no other alleles of IGHV_S1 were identified in the genotype: the exclusive association with *03 therefore provides additional support for the novel inference.