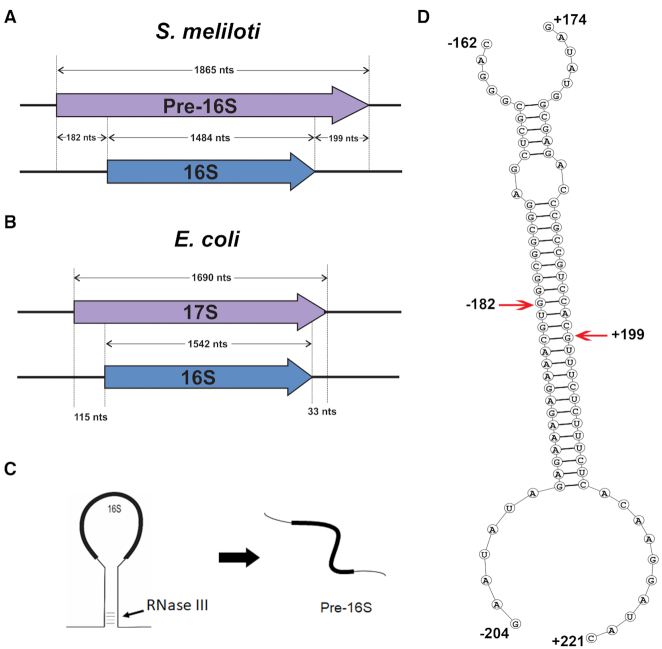
Figure 6.
Characterization of the pre-16S band. (A) The pre-16S RNA was isolated by gel excision and the RNA ends were determined using the RACE protocol described in Materials and methods and FIG S8. Locations of the pre-16S (purple) ends are represented in comparison with the mature 16S rRNA (blue). (B) Length of the 17S rRNA (purple) relative to 16S (blue) in E. coli is shown. Size of the 17S in E. coli is relatively smaller compared to the pre-16S of S. meliloti. (C) A simplified view of RNase III cleavage of the rRNA primary transcript. RNase III cleaves the double-stranded stem structure formed by the nascent primary rRNA transcript thereby releasing the pre-16S RNA. (D) Secondary structure of the rRNA region with the pre-16S ends indicated with red arrows. Analysis of the ends reveals that they originated from the RNA regions that are complementary to each other and are capable of forming a 31 bp stem with two mismatches in between.