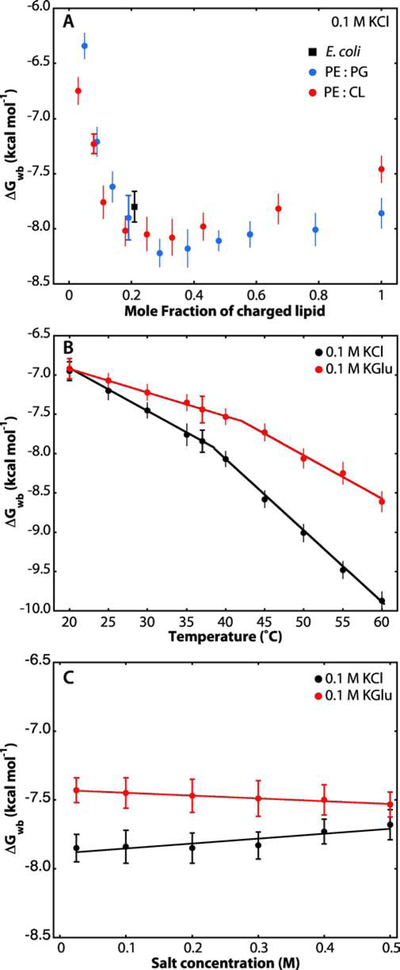
Figure 3. LUV lipid composition, temperature, and electrolyte concentration affect the partitioning of SecA into LUVs.
Data points with horizontal caps represent the SEMs for experiments performed in triplicate. Data points lacking horizontal caps indicate single titration experiments. In that case, the vertical error bars represent estimates of uncertainties determined from non-linear least-squares fits. (A) Negatively charged lipids (POPG or CL) strongly improve SecA partitioning into lipid vesicles, as established earlier by other laboratories [9, 12]. Free energies of transfer from water to bilayer were determined from the partitioning of SecA at 37°C into LUVs containing an increasing fraction of POPG (blue) or cardiolipin (red) monitored by Trp fluorescence. As a reference, partitioning into LUV formed from E. coli lipids is also plotted (black square). (B) Influence of temperature on the partitioning of SecA to liposomes made from E. coli lipids in the presence of 0.1 M KCl (black) or 0.1 M KGlu (red). The increases in the magnitudes of ΔGwb with temperature are likely due to partial thermal unfolding of SecA, which leads to exposure of otherwise buried hydrophobic residues. The temperature effect is smaller in KGlu compared to KCl, as expected from the stabilization of SecA by glutamate [45]. (C) Effect of electrolyte concentration on free energies of transfer of SecA at 37 °C. The free energies in KGlu are slightly lower than in KCl, likely due to glutamate stabilization that reduces exposure of normally-buried hydrophobic residues [45]. Linear fits to the data of panel C are shown. It remains to be determined why the straight-line fits to the points have opposite slopes and why they converge to ΔGwb = −7.6 kcal mol−1 at 0.8 M salt concentration. Linear-fit parameters: KCl, intercept at 0 mM salt is −7.9 with a slope of +0.36. KGlu, intercept at 0 mM salt is −7.4 with a slope of −0.20