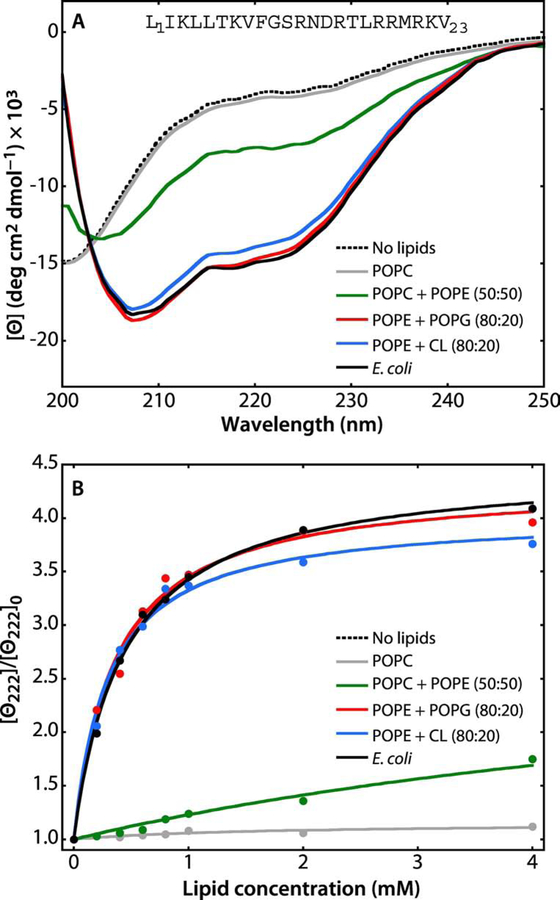
Figure 5. A synthetic peptide corresponding to the N-terminal 23 residues of SecA partition strongly into LUV.
The N-terminal domain of SecA has long been known to be critical for the interaction of SecA with lipids [54, 55] and it has been shown that the first ten residues form an amphipathic helix in the membrane interface [16]. Because the peptide contains no Trp residue, partitioning was determined by measuring molar ellipticity at 222 nm. The general behavior of the peptide in the absence and presence of lipid vesicles is very similar the behavior of the melittin 26-residue amphiphilic peptide [56–58]. For all measurements, the peptide concentration was 40 μM. (A) In the absence of LUV (dotted black curve) or in the presence POPC LUVs (gray curve), the spectra are those expected for unfolded/unbound peptides. There is weak binding of the peptide to POPC:POPE (50:50) LUV (green curve), indicated by the appearance of a weak α-helical signal at 222 nm. The α-helicity of the peptide increases dramatically in the presence of POPE:POPG (80:20, red curve), POPE:CL (80:20, blue curve), or E. coli lipids (black curve). (B) Titration curves obtained by titrating the 40 μM peptide solution with LUVs of varying composition at 37 °C monitored by the change in molar ellipticity ([Θ]) at 222 nm. The color codes are the same as in panel A. The titration curves show that the peptide binds most strongly to E. coli lipids. The partition coefficients (Kx) and free energies of transfer (ΔGwb) determined from these curves are shown in Table 2.