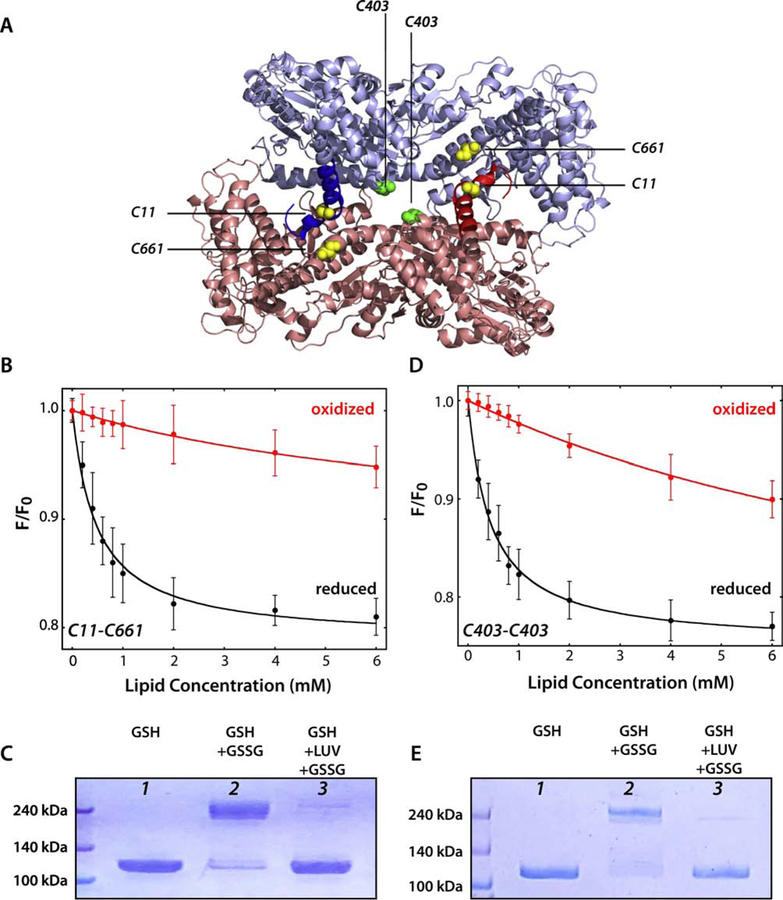
Figure 6. SecA binds weakly to LUV as the 1M6N dimer.
Cysteines were engineered into a Cys-free SecA construct to stabilize the 1M6N dimer under oxidizing conditions. (A) 3D structure of the 1M6N dimer in a head-to-tail disposition. The stabilizing pairs of cysteines for mutant C11-C661 and C403 are highlighted in yellow and green, respectively. (B) Titration with E. coli LUVs of the C11-C661 dimer (4 µM) under oxidizing (red curve) or reducing conditions (black curve). The Cys-stabilized dimer binds very weakly compared to the monomer (Table 3). (C) Coomassie blue-stained gels of C11–661-SecA protein under different conditions. Lanes 1 and 2 show, respectively, that SecA is monomeric under reducing conditions and dimeric under oxidizing conditions. For lane 2, an excess of oxidized glutathione was introduced by dialysis. In lane 3, SecA was incubated with LUV (E. coli lipids, 4 mM) for 30 minutes under reducing conditions to allow equilibrium binding of ~95% of SecA. Then, oxidized glutathione was introduced by dialysis to cross link any SecA dimers that existed on the membrane. The great preponderance of SecA migrated on the gel as a monomer, indicating that SecA either partitioned as a monomer or as a dimer different from 1M6N formed upon partitioning. (D) Titration of C403 SecA with E. coli LUVs under oxidizing (red curve) or reducing conditions (black curve). The Cys-stabilized dimer binds very weakly compared to the monomer (Table 3). (E) Coomassie blue-stained gels of C403-SecA protein using the same protocol as in panel C. The results are consistent with membrane-bound SecA being monomeric, as in panel C.