Figure 7.
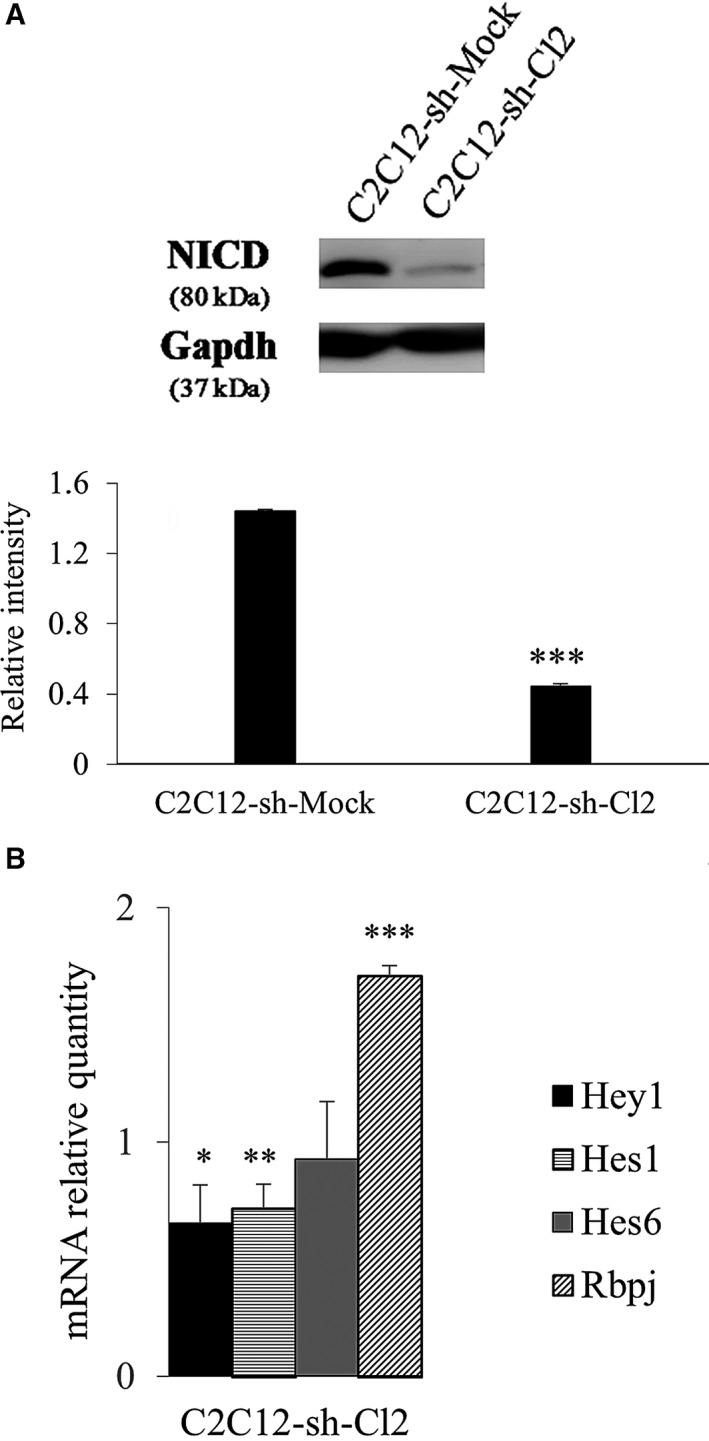
The expression of target genes of Notch, Hey1 and Hes1, decreased in st6gal1‐knockdown cells. (A) Relative amounts of NICD (the active form of Notch which binds to Rbpj in order to transcriptionally activate target genes as Hes1 and Hey1) determined by western blot using total proteins from proliferating C2C12‐sh‐Mock and C2C12‐sh‐Cl2 cells. Protein levels were determined in comparison with gapdh loading control. This experiment has been performed three times (n = 3). (B) Expression of Notch target genes Hey1 and Hes1 (encoding transcriptional repressors regulating myogenesis), Hes6 whose expression is not under the control of Notch signaling (encoding an independent protein of the Notch signaling pathway) and Rbpj (encoding a transcription factor) in C2C12‐sh‐Mock and C2C12‐sh‐Cl2 cells in proliferation. Fold changes are expressed relative to C2C12‐sh‐Mock cells. Data are expressed as relative transcript amounts corresponding to the ratio of C t values for C2C12‐sh‐Cl2 cells and C t values for C2C12‐sh‐Mock cells and normalized against the expression of gapdh transcript. Statistical analyses of gene expression were performed by comparison with the C2C12‐sh‐Mock (which was set as 1) for three independent experiments (n = 3). Means ± SEMs are shown (two‐tailed t‐test, with a significant level of *P < 0.05; **P < 0.01; ***P < 0.001).
