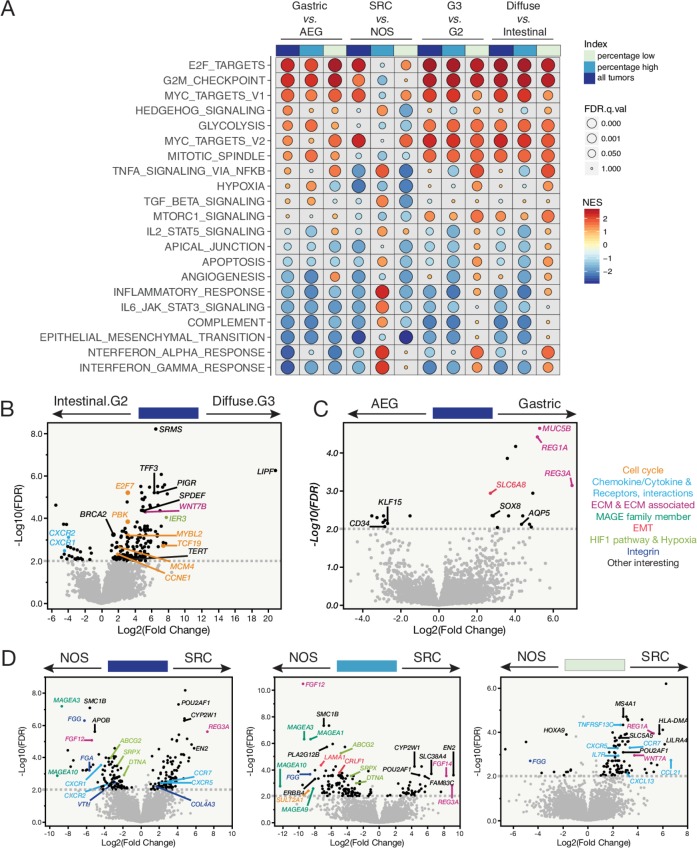
Figure 4.
Transcriptome profiling of peritoneal carcinomatosis (PC) specimens—enriched signalling pathways and differentially expressed genes (DEGs) associated with disease aggressiveness. (A) GSEA pathway analysis showing enriched signalling pathways by histological subtypes (eg, gastric vs adenocarcinoma of esophageal and gastric junction (AEG)) for all PC specimens together, PC specimens with higher tumour cell percentages and those with lower tumour cell percentages, respectively (top track). Each row represents a MSigDB Hallmark pathway. The size of the filled circle reversely reflects the FDR q values, the bigger the circle the smaller the q value. NES, normalised enrichment score. The colour tone of NES indicates the direction of enrichment, with warm colour denotes positive enrichments and cold colour denotes negative enrichment. (B) The DEGs identified in G3 diffuse against G2 intestinal subtypes. Dashed line indicates a cut-off of FDR q value≤0.01. Genes that are above the dashed line with a fold change≥2, cancer related or biologically important are labelled by their names in the volcano plot. (C) The DEGs identified in gastric against AEG subtypes of PC specimens. (D) The DEGs identified in signet-ring cell (SRC) vs NOS subtypes in all PC specimens together (left), PC specimens with higher (middle), or lower tumour cell percentage (right), respectively.