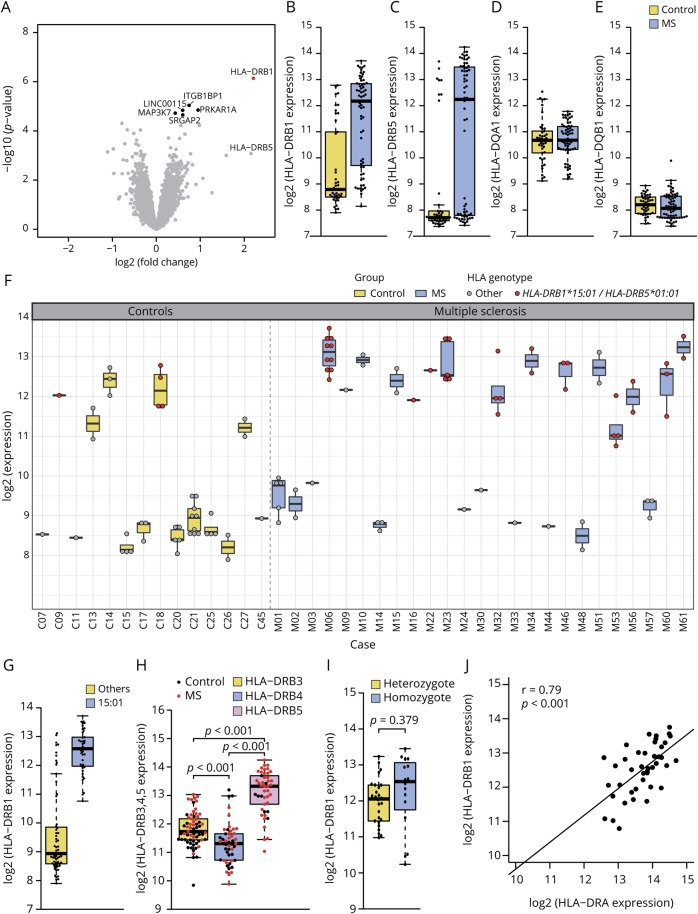
Figure 2. Differential gene expression in MS NAGM vs control case cortical gray matter.
(A) Volcano plot of the differential gene expression analysis between MS NAGM and control gray matter (GM) revealed HLA-DRB1 as the most significant differentially regulated gene (FC = 4.62, adj. p value = 0.013, marked in red). Differentially expressed genes with an adjusted p value between 0.05 and 0.1 are marked in black. This was the case for ITGB1BP1 (FC = 1.67, adj. p value = 0.065), PRKAR1A (FC = 1.93, adj. p value = 0.065), LINC00115 (FC = 1.5, adj. p value = 0.065), MAP3K7 (FC = 1.34, adj. p value = 0.067), and SRGAP2 (FC = 1.5, adj. p value = 0.067). Boxplots show the log2 gene expression of HLA-DRB1 (B), HLA-DRB5 (C), and HLA-DQA1 (D) and HLA-DQB1 (E) between MS NAGM and GM. HLA-DRB1 is significantly differentially expressed between MS NAGM and GM (B). Both, HLA-DRB1 (B) and HLA-DRB5 (C) show a bimodal expression pattern in MS and control tissue, whereas the expression of HLA-DQA1 (D) was normally distributed within the sample groups, and HLA-DQB1 (E) was at the lower detection limit. (F) Boxplots show log2 of the HLA-DRB1 gene expression in all tissue samples and in all cases used for the microarray analysis. All cases with the HLA-DRB1*15:01 genotype show a high HLA-DRB1 expression (red dots). (G) HLA-DRB1 expression between samples from cases carrying the HLA-DRB1*15:01 or other HLA-DRB1 alleles. Boxplot shows that all HLA-DRB1*15:01 tissue samples belong to the HLA-DRB1 high expresser group. (H) HLA-DRB3, 4, and 5 expression of all samples. (I) Comparison of hetero- and homozygote carriers of the HLA-DRB1*15:01 allele. (J) Correlation of HLA-DRB1 with HLA-DRA within the HLA-DRB1*15:01 positive samples. (H and I) p Values are derived from a 2-sided Welch t test. (J) p Value is derived from a linear model.