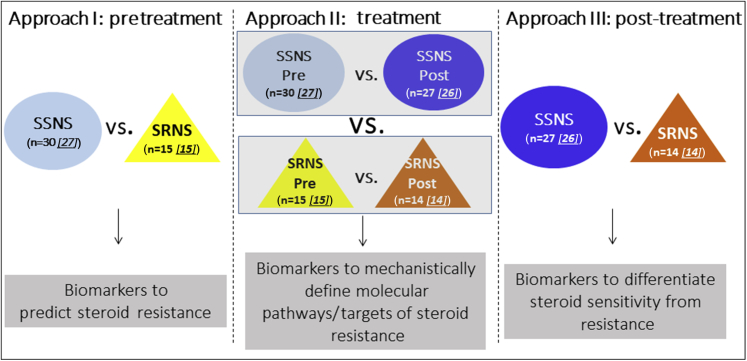
Figure 1.
Study hypothesis and design. The present studies were designed to test the hypothesis that metabolomic analyses of paired plasma samples from children with steroid-sensitive nephrotic syndrome (SSNS) and steroid-resistant nephrotic syndrome (SRNS) can be used to identify biomarkers able to (approach I) predict clinical steroid resistance, (approach II) define specific mechanistic molecular pathways or targets associated with clinical steroid resistance, and (approach III) differentiate clinical steroid sensitivity from steroid resistance. SSNS Pre, light blue circle; SRNS Pre, yellow triangle; SSNS Post, dark blue circle; SRNS Post, orange triangle. Starting patient samples: n = 86 (SSNS Pre = 30, SSNS Post = 27, SRNS Pre = 15, SRNS Post = 14). Approach I: Thirty SSNS patient samples were subjected to 1H NMR and 27 were analyzed after removing 2 outliers and 1 steroid-dependent [SD] patient; 15 SRNS patient samples were subjected to HNMR and were analyzed. Approach II: Twenty-seven SSNS pre-patient samples were analyzed after removing 2 outliers and 1 SD patient; 26 SSNS post-patient samples were analyzed after removing 1 SD patient; 24 sample pairs were analyzed for relative concentration data; 15 SRNS pre-patient samples were analyzed; 14 SRNS post-patient samples were analyzed; and 14 pairs were analyzed for relative concentration data. Approach III: Twenty-six SSNS post-patient samples were analyzed after removing 1 SD patient; 14 SRNS post-patient samples were analyzed.