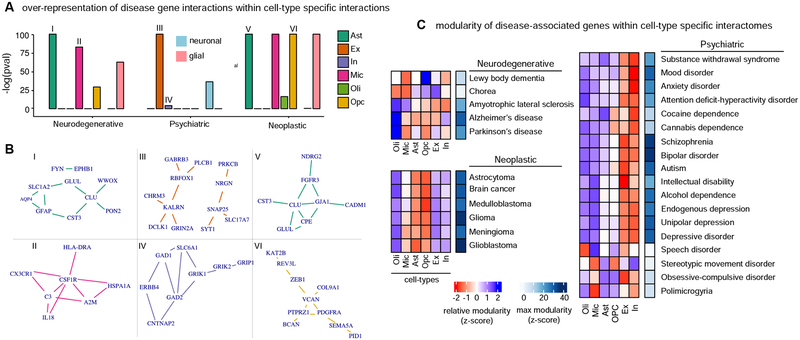
Figure 4. Brain-associated disorders converge to cell-type specific modular perturbations,
a, over-representation scores for gene interactions between disease-associated genes within cell-type specific interactions are shown for neurodegenerative, psychiatric, and neoplastic disorder groups. Cell-type (Ast, In, Oli, Ex, Mic, OPC) and cell-type group (neuronal, glial) over-representation analysis are considered. Neuronal (blue) or glial (pink) over-representation is highlighted with a rectangle. Roman numbers link cell-type and disease interaction sets with corresponding networks in b. b, top 10 strongest interactions for to associated interaction sets considering in a are shows as networks, c, network modularity scores for genes associated with individual diseases across cell-types are shown. Modularity is estimated by a compactness score that measures the deviation of module size (average measure of inter-gene network distance) relative to random expectation. Relative (z-scaled) modularity across cell-types is shown in blue-red scale. Maximal compactness for a disease is shown in blue scale. See also Figure S5.