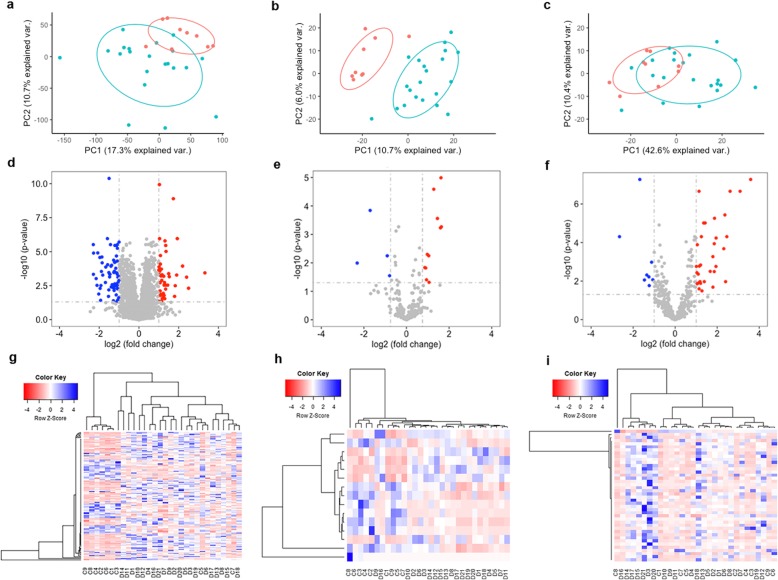
Fig. 3.
Differential gene expression analysis. a–c Principal component analysis (PCA) of mRNAs (a), lncRNAs (b), and microRNAs (miRNAs) (c), respectively. Each dot represents one sample. Blue: heart failure (HF) samples. Red: control samples. d–f Volcano plots showing mRNA (d), lncRNA (e), and miRNA (f) differential expression. Red and blue dots denote significantly upregulated and downregulated genes, respectively. g–i Heatmap plots showing differentially expressed mRNAs, lncRNAs, and miRNAs among the samples. X-axis: sample IDs starting with a D denote HF samples and samples with a C for control samples