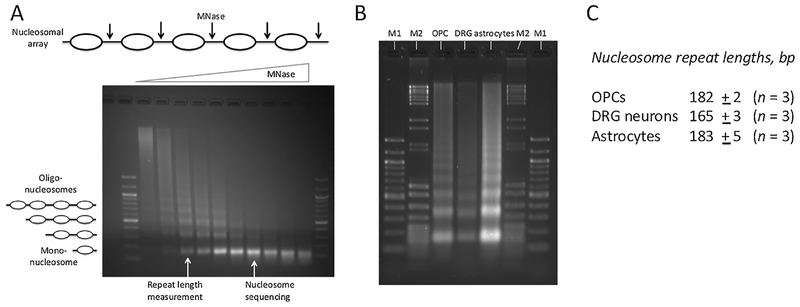
Fig. 1.
DRG neurons have shorter nucleosome spacing than OPCs and astrocytes. (A) Gel electrophoresis of DNA purified from a typical MNase titration (example: astrocytes). The oligo-nucleosomes become progressively shorter as more linkers are cut by MNase, eventually resulting in mono-nucleosomes. The repeat length is measured using samples with multiple nucleosome bands (as indicated). Nucleosome positioning (MNase-seq) data are obtained using samples containing predominantly mono-nucleosomes, avoiding over-digestion (indicated by the smearing below the mono-nucleosome band in the last 3 samples). 100-bp marker. (B) Typical gel used for repeat length measurement. Specific samples from titrations like that in A are analyzed side-by-side. M1: 100-bp marker; M2: mixture of pBR322 MspI and λ-DNA BstEII digests. (C) Repeat length data (average of 3 independent experiments with standard deviation). The lengths of the mono-, di-, tri- and tetra-nucleosomes were determined using the markers for calibration. The repeat length is given by the slope of the regression line in a plot of DNA length vs. nucleosome number (Fig. S5).