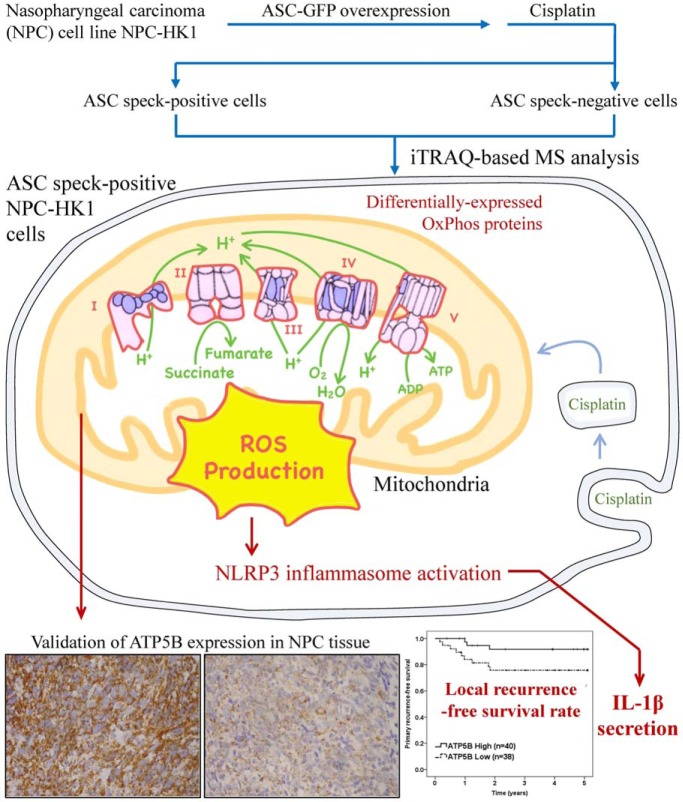
Using flow cytometry analysis, followed by iTRAQ technology, ASC speck(+) cells were discovered to be enriched with mitochondrial OxPhos proteins, which modulate mtROS production and downstream inflammasome activation. Two proteins, NDUFB8 (complex I) and ATP5B (complex V) are found associated with local recurrence-free survival in NPC patients. Multivariate analysis additionally indicated that NDUFB8 and ATP5B may serve as markers for local recurrence. This study experimentally links mitochondrial OxPhos proteins, inflammasome activation and clinical outcomes among NPC patients.
Keywords: iTRAQ, inflammatory response, mitochondria function or biology, oxidative stress, cancer biology, nasopharyngeal carcinoma
Graphical Abstract
Highlights
Flow cytometry analysis is used to isolate ASC speck(+) NPC cells.
Proteome analysis of ASC speck(+) NPC cells reveals enriched mitochondrial OxPhos proteins.
OxPhos proteins mediate NLRP3 inflammasome activation through mtROS.
OxPhos proteins, NDUFB8 and ATP5B are correlated with NPC local recurrence.
Abstract
We previously reported that tumor inflammasomes play a key role in tumor control and act as favorable prognostic markers in nasopharyngeal carcinoma (NPC). Activated inflammasomes frequently form distinguishable specks and govern the cellular secretion of IL-1β. However, we know little about the biological and biochemical differences between cells with and without apoptosis-associated speck-like protein containing a caspase-recruitment domain (ASC) speck formation. In this study, we used proteomic iTRAQ analysis to analyze the proteomes of NPC cells that differ in their ASC speck formation upon cisplatin treatment. We identified proteins that were differentially over-expressed in cells with specks, and found that they fell into two Gene ontology (GO) pathways: mitochondrial oxidative phosphorylation (OxPhos) and ubiquinone metabolism. We observed up-regulation of various components of the OxPhos machinery (including NDUFB3, NDUFB8 and ATP5B), and subsequently found that these changes lead to mitochondrial ROS (mtROS) production, which promotes the formation and activation of NLRP3 inflammasomes and subsequent pyroptosis. In NPC patients, better local recurrence-free survival was significantly associated with high-level expression of NDUFB8 (p = 0.037) and ATP5B (p = 0.029), as examined using immunohistochemistry. However, there were no significant associations between the expression of NDUFB8 and ATP5B with overall survival of NPC patients. Together, our results demonstrate that up-regulated mitochondrial OxPhos components are strongly associated with NLRP3 inflammasome activation in NPC. Our findings further suggest that high-level expression of OxPhos components could be markers for local recurrence and/or promising therapeutic targets in patients with NPC.
Inflammation plays important roles at different stages of tumorigenesis. A key signal involved in driving acute and chronic inflammation is controlled by the inflammasome, which is an inducible cytoplasmic multiprotein complex that serves as a platform for sensing danger signals in the tumor microenvironment (1, 2). Various inflammasomes have been identified. Some comprise intracellular pattern recognition receptors, such as nod-like receptors and AIM2-like receptors, which can be activated by pathogen-associated molecule patterns and damage-associated molecule patterns (3). The apoptosis-associated speck-like protein containing a caspase-recruitment domain (ASC)1 protein, also called PYD and CARD domain containing (PYCARD), is encoded by PYCARD gene in human and is a key component of all known inflammasomes. Upon initiation of inflammasome activation, all inflammasomes recruit the adaptor protein ASC assembles into a large helical fibril called the ASC speck (4). The ASC speck serves as signaling platform for caspase-1, leading to the activation of caspase-1, the maturation of pro-inflammatory cytokines (e.g. IL-1β) and the initiation of a lytic form of programmed cell death known as pyroptosis (5).
Nasopharyngeal carcinoma (NPC) is a cancer that is prominent in the Taiwanese population. It is closely associated with Epstein-Barr virus (EBV) infection. In tumor cells, EBV-encoded latent membrane protein 1 (LMP1) mediates pro-IL-1β gene expression through NF-κB signaling (6). Within the NPC tumor mass or tumor microenvironment, the levels of proinflammatory cytokines, including IL-1β, are elevated (7). Indeed, elevated levels of IL-1β and the NLRP3, AIM2 and RIG-I inflammasomes were reported to be useful as prognostic biomarkers for predicting better recurrence-free survival of NPC patients treated with the current protocols (6). End-binding protein 1 (EB1), which regulates microtubule polymerization, was reported to co-immunoprecipitate with ASC of the NLRP3, AIM2, and RIG-I inflammasomes in NPC cells and was found to be required for speck formation in response to AIM2 inflammasome activation (8). EB1 has also been shown to mediate the autophagy-dependent secretion of IL-1β, and the EB1-mediated, autophagy-based and inflammasome-induced secretion of IL-1β was found to be regulated by 5′AMP activated protein kinase (9). Other studies found that the NLRP3 inflammasome is activated by EBV-LMP1 and the chemotherapeutic agent, cisplatin, in NLRP3-knockdown NPC cells (6, 10). However, we know relatively little about the changes in other cellular components of activated inflammasomes.
Emerging evidence indicates that mitochondria not only act as the energy powerhouses of the cell, they also function as signaling organelles that can activate signal pathways to regulate innate and adaptive immunity (11). Recent studies have indicated that mitochondrial dysfunctions may crucially modulate NLRP3 inflammasome activation (12). NLRP3 inflammasome activation involves various parameters that can be altered under mitochondrial dysfunction, such as mitochondrial membrane potential (mtΔΨ) (13), mitochondrial outer membrane permeabilization (14), and the levels of mitochondrion-derived molecules [(e.g. mitochondrial ROS (mtROS) (15), release of mitochondrial DNA (mtDNA) (16), and cardiolipin (17)] and mitochondrion-residing molecules [(e.g. mitochondrial antiviral signaling protein (MAVS) (18) and mitofusin 2 (Mfn2) (19)]. Notably, MAVS and Mfn2 help redistribute cytosolic NLRP3 to mitochondrial outer membrane and enhance NLRP3 inflammasome activation. Further, mtROS can oxidize mtDNA, leading to further NLRP3 inflammasome activation.
The electron transport chain (ETC) is in the mitochondrial inner membrane. It is the major source of oxidative phosphorylation (OxPhos) and is involved in the generation of energy: oxygen acts as an electron acceptor, and electrons from NADH and FADH2 generated by glycolysis and the TCA cycle are used to generate ATP. Through the ETC, protons are transported across the inner membrane of mitochondria to generate mtΔΨ. Mechanistically, the loss of mitochondrial potential has been associated with the production of mtROS (15) and changes in cellular Ca2+ (20) in the context of NLRP3 inflammasome activation. Moreover, mitochondria are the major source of cellular ROS (21). Several lines of evidence support the notion that mtROS activates NLRP3 inflammasome formation and activation. For example, inhibitors of OxPhos trigger mtROS production, which can lead to NLRP3 inflammasome activation (15). Meanwhile, we previously showed that the E3 ubiquitin ligase, Cbl, maintains mitochondrial size and reduces mtROS production by suppressing NLRP3 inflammasome activation (22). Together, the previous results reveal that mtROS can be an upstream regulator for NLRP3 inflammasome activation.
Here, we report that mitochondrial OxPhos components are enriched in ASC speck-positive NPC cells and are likely to participate in NLRP3 inflammasome activation and cell pyroptosis. Further, the components of OxPhos are highly expressed in tumor cells and significantly correlated with better post-treatment survival of NPC patients. Our findings further clarify the molecular mechanisms of mitochondria-mediated NLRP3 inflammasome activation and suggest that overexpressed OxPhos proteins could prove useful as favorable markers for local recurrence in NPC patients.
EXPERIMENTAL PROCEDURES
Cell Culture
EBV-positive NPC cell line, NPC-HK1-EBV, and an EBV-negative NPC cell line, NPC-HK1, were kindly provided by Dr. S. W. Tsao (Hong Kong University, SAR, China) and cultured as previously described (6, 23). The NPC-BM1 cell line was established from a bone marrow biopsy of a female Taiwanese patient with NPC and cultured as previously described (24). For generation of NPC-HK1-ASC-GFP and NPC-BM1-ASC-GFP cells, the NPC-HK1 and NPC-BM1 cells were transduced with lentiviral particles containing an ASC-GFP plasmid, using a commercial lentivirus transduction protocol (Sigma-Aldrich, St. Louis, MO). For inflammasome activation, the cells were treated with 40 μm cisplatin (cat# P4393; Sigma-Aldrich), 10 μm nigericin (cat# N7143; Sigma-Aldrich), 5 mm ATP (cat# A7699; Sigma-Aldrich), 200 μm monosodium urate (MSU) crystals (cat# tlrl-msu; InvivoGen, San Diego, CA) or 200 μm Alum (cat# tlrl-alk; InvivoGen). For mtROS scavenger treatment, the cells were preincubated with MitoTEMPO (cat# SML0737; Sigma-Aldrich) for 1 h.
Generation and flow cytometric isolation of ASC-GFP speck-forming cells
NPC-HK1-ASC-GFP and NPC-BM1-ASC-GFP cells were treated with 40 μm cisplatin for 24 h (NPC-HK1-ASC-GFP) or 30 h (NPC-BM1-ASC-GFP), and ASC speck(+) and speck(−) cells were isolated by flow cytometry as previously described (22, 25). Briefly, the transit of ASC-GFP protein into the speck is detected by a decreased pulse width (W) of emitted fluorescence after the inflammasome activation, and the total amount of ASC-GFP stained per cell (pulse area, A) was higher in cells with ASC speck. Therefore, low and high pulse width to pulse area profile (ASC-W:ASC-A profile) were taken as indicating ASC speck(+) cells and ASC speck(−) cells, respectively). We then sorted for the presence or absence of ASC-GFP specks by flow cytometric method.
Sample Preparation for Tryptic Digestion and iTRAQ Labeling
NPC-HK1-ASC-GFP and NPC-BM1-ASC-GFP cells were isolated by flow cytometry and lysed in buffer containing 100 mm triethylammonium bicarbonate (TEABC; Sigma-Aldrich) and 0.1% RapiGestTM SF (Waters Corporation, Milford, MA) on ice for 15 min. The cell lysate was collected, sonicated on ice and centrifuged at 10,000 × g for 25 min at 4 °C. The resulting supernatant was used as the cell extract. Protein concentrations were determined using a Pierce BCA protein assay kit (Thermo Fisher Scientific, San Jose, CA). For tryptic in-solution digestion, 20 μg protein of each sample was reduced with 4.8 mm Tris-(2-carboxyethyl)-phosphine (Sigma-Aldrich) and 260 mm TEABC at 60 °C for 1 h and then alkylated with 9.6 mm methyl methanethiosulfonate (Sigma-Aldrich) at room temperature for 30 min. The proteins were digested at 37 °C overnight with modified sequencing-grade porcine trypsin (Promega, Madison, WI) using a trypsin-to-protein ratio of 1:1 (w/w). Trypsin-digested peptides were labeled with iTRAQ reagents (AB Sciex, Foster City, CA) according to the manufacturer's protocol. The peptides from ASC speck(−) and speck(+) cells were labeled with iTRAQ 114 and 115 tags, respectively. For the biological replicates, the iTRAQ 116 and 117 tags were incubated with peptide samples obtained from different batches of ASC speck(−) and speck(+) cells. After incubation for 1 h at room temperature, the four labeled samples were pooled, frozen and lyophilized to dryness using a Proteomic CentriVap Concentrator System (Labconco, Kansas City, MO).
The peptides were desalted using a ziptip (Merck Millipore, Billerica, MA) packed with Resin-C18 (GE Healthcare, UK). In brief, a sufficient volume of Resin-C18 (1 μl for 10 μg of proteins) was packed into a ziptip. To activate the Resin-C18, the ziptip was sequentially loaded with 200 μl of acetonitrile (Mallinckrodt Baker, Ireland) at concentrations of 75%, 40 and 3% in 1% trifluoroacetic acid (TFA), and then centrifuged at 300 rpm for 30 s. This step was repeated three times to ensure complete activation. Dried iTRAQ-labeled peptide samples were reconstituted with 0.1% TFA to a final protein concentration of 1 μg/μl. The reconstituted peptides were loaded into the ziptip, centrifuged at 300 rpm for 30 s and washed with 3% acetonitrile in 1% TFA. Finally, the iTRAQ-labeled peptide samples were eluted from the ziptip with 30%, 50%, and 70% acetonitrile in 1% TFA and dried by vacuum centrifugation.
Peptide Fractionation and LC-MS/MS Analysis
The iTRAQ-labeled peptides were separated using an on-line 2D-HPLC system (Dionex Ultimate 3000, Thermo Fisher Scientific) and analyzed as previously described (26). Briefly, peptides (20 μg) were resuspended in 0.1% formic acid (20 μl) and desalted using a ziptip home-packed with C18 resin (5–20 μm, LiChroprep RP-18; Merck, Taipei, Taiwan). The resultant samples were vacuum-dried, reconstituted in 50 μl of buffer A (0.1% formic acid and 30% acetonitrile), and loaded onto the homemade column (Luna SCX, 5 μm, 0.5 × 180 mm) at a flow rate of 5 μl/min for 30 min. The peptides were then eluted with a 0–100% gradient of buffer B (0.5 m ammonium chloride, 30% acetonitrile and 0.1% formic acid). The resulting 44 peptide fractions were diluted in-line prior to being trapped onto the Zorbax 300SB-C18 column (0.3 × 5 mm; Agilent Technologies, Wilmington, DE). Each fraction was then separated on a homemade column (HydroRP 2.5 μm, 75 μm inner diameter and 20 cm length) with a 15-μm tip using buffer C (acetonitrile containing 0.1% formic acid). A linear gradient of buffer C (3–28% for 37 min, 28–50% for 12 min, 50–95% for 2 min, 95% for 5 min and 3% for 9 min) was applied at a flow rate of 0.3 μl/min.
The LC equipment was connected to the LTQ-Orbitrap ELITE mass spectrometer (Thermo Fisher Scientific), which was operated using the Xcalibur software (Version 2.2, Thermo Fisher Scientific). Full-scan MS was performed in the Orbitrap over a range of 400 to 2000 Da and a resolution of 60,000 at m/z 400. The ion signals of (Si(CH3)2O)6H+ at m/z 445.120025, 462.146574 and 536.165365 were used for lock masses and internal calibration. Each 12 data-dependent MS/MS scan events of six collision-induced dissociations (CID) and six higher-energy collision-induced dissociations (HCD) were followed by one MS scan for the six most abundant ions in the preview MS scan. The m/z values selected for MS2 were excluded dynamically for 40 s with a relative mass window of 1.5 Da. The electrospray voltage was set to 1.8 kV and the temperature of the capillary was set to 220 °C. Automatic gain control was applied to preclude over-filling of the ion trap, and 1000 ms/2 × 106 ions, 150 ms/5 × 103 ions and 300 ms/3 × 104 ions were set as the maximum accumulated time/ions for the full scan, CID and HCD, respectively.
Sequence Database Searching and Quantitative Data Analysis
Protein database searching was performed using the Proteome Discoverer software (version 1.4; Thermo Fisher Scientific) with the reporter ion quantifier node for iTRAQ data analysis. The MS/MS spectra were searched against the Swiss-Prot human sequence database (released 2018/03, selected for Homo sapiens; 20,198 entries) using the Mascot search engine (version 2.2.0; Matrix Science, London, UK). For peptide identification, one missing tryptic cleavage was allowed. Mass tolerances of 10 ppm, 0.5 Da and 0.05 Da were permitted for intact peptide masses, CID fragment ions and HCD fragment ions, respectively. Oxidized methionine (+16 Da) was considered a potential variable modification, and iTRAQ (N-terminal, +144 Da), iTRAQ (K, +144 Da) and methyl methanethiosulfonate (C, +46 Da) were considered fixed modifications. The search results (peptide-spectrum matches) were filtered for a highly confident peptide identification in Proteome Discoverer to ensure that the overall false discovery rate was less than 0.01. Identification of epithelial keratins was excluded. Proteins with single peptide hits were removed. Each quantified protein contained at least two quantifiable spectra. The quantitative data were exported from Proteome Discoverer and manually normalized such that the log2 of protein ratios displayed a median value of zero for all peptides in each protein. This was performed across an entire labeling experiment to correct for any variation in protein abundance. The cutoff value for determining whether a protein was considered dysregulated was selected according to a comparison of protein levels between the same ASC speck(−) samples in two different batches. Proteins with a log2 ratio greater than the mean ratio plus one standard deviation (S.D.) and those with a log2 ratio less than the mean ratio minus one S.D. were overexpressed and underexpressed, respectively.
RNA Interference
NDUFB3 (sense 5′-UGGCU UUGCA AAGAG UGUUU C-3′, 5′- CCGCA AUGAA GCUUG GAGAU A-3′), NDUFB8 (sense 5′-CCAAAG CAGUA UCCUU ACAAU-3′, 5′-CCUGG CUUUC AUGAU AUUCA U-3′), NDUFB5 (sense 5′- GUCAA GCUGA ACUAG CAGAA A-3′, 5′-CGAAA GCAAC UCCUG ACAAU U-3′), MT-CO2 (sense 5′-CCAUC AUCCU AGUCC UCAU-3′, 5′-GAUCC CUCCC UUACC AUCAA A-3′, 5′-GCAAU UCCCG GACGU CUAA-3′), ATP5B (sense 5′-GCACA GUAAG GACUA UUGCU A-3′, 5′-GCGUU UCUUG UCUCA GCCAU U-3′), ATP5H (sense 5′- CCCGU GCCAG AGGAU AAAUA U-3′, 5′-CCAUU GCUAG UUCCC UGAAA U-3′) and ATP5J (sense 5′- GAUCC UAUAC AGAAA CUCUU U-3′, 5′-GAGGA CCUGU UGAUG CUAGU U-3′) siRNAs were purchased from MDBio (Taipei, Taiwan). Negative-control siRNA were purchased from Invitrogen (Carlsbad, CA). Cells were transfected with 50 nm dsRNA duplexes using jetPRIME (Polyplus Transfection, New York, NY) according to the manufacturer's instructions. After 4 h of incubation, the dsRNA complexes were removed, and the cells were re-plated in 5 ml of fresh culture medium.
Western Blotting
Western blot analysis was performed as described previously (22). Briefly, cells were lysed in RIPA buffer containing a protease inhibitor mixture (cat# 04693116001; Roche Applied Science, Indianapolis, IN) on ice for 30 min, followed by centrifugation (12,000 rpm for 10 min at 4 °C). The protein lysates were further resolved by SDS-PAGE and transferred to nitrocellulose membranes (Amersham Biosciences, UK). The membranes were incubated with primary anti-human antibodies against NDUFB8 (1:1000; cat# HPA003886; Sigma-Aldrich), ATP5D (1:2000; cat# A9929; ABclonal, Woburn, MA), ATP5J (1:2000; cat# A3751; ABclonal), ATP5H (1:2000; cat# A4425; ABclonal), NDUFB3 (1:500; cat# SC-393351; Santa Cruz Biotechnology, Santa Cruz, CA), DNUFB5 (1:1000; cat# SC-514245; Santa Cruz Biotechnology), ATP5B (1:1000; cat# SC-55597; Santa Cruz Biotechnology), ATP5F1 (1:500; cat# sc-514419; Santa Cruz Biotechnology), GAPDH (1:2000; cat# SC-32233; Santa Cruz Biotechnology), actin (1:3000; cat# MAB1501; Merck Millipore), or MT-CO2 (1:2000; cat# A6404; Invitrogen). The blots were reacted with an HRP-conjugated secondary antibody (1:5000; Sigma-Aldrich) and the immunoreactive bands were detected with the TOOLS Extreme ECL-HRP Substrate (cat# TU-ECL03; Biotools, Taiwan).
IL-1β ELISA
Human IL-1β in cell culture supernatants was detected using the Human IL-1β ELISA Ready-SET-Go Kit (cat# 88–7261; eBioscience, San Diego, CA) according to the manufacturer's instructions.
MtROS Measurement
MtROS were measured using MitoSox (cat# M36008, Invitrogen) according to the manufacturer's protocol and as previously described (22). The NPC cells were incubated with MitoSox (5 μm) at 37 °C for 20 min and then analyzed by flow cytometry.
Lactate Dehydrogenase (LDH) Release Assay
LDH levels were detected in the culture supernatant using an LDH Cytotoxicity detection kit (Takara Bio Inc., Shiga, Japan) according to the manufacturer's instructions and as previously described (27). Briefly, each culture supernatant was collected, centrifuged at 1200 rpm for 5 min at 4 °C and incubated with substrate. The absorbance of the red formazan product was measured at 490 nm using a SpectraMax M2 microplate reader (Molecular Devices, San Jose, CA). The formula for calculating the percentage of LDH release was as follows: [(LDH sample)-(LDH negative control)]/[(LDH positive control)-(LDH negative control)]. The LDH spontaneously release from untreated cells served as the LDH negative control, whereas the maximum LDH released from 2% Triton-X-100-treated cells served as the LDH positive control.
Determination of Caspase-1 Activity and Pyroptotic Cell Death
Active caspase-1 was detected using the Fluorochrome Labeled Inhibitors of Caspase-1 (FLICA, FAM-YVAD-FMK) reagent (Immunochemistry Technologies, Bloomington, MN) according to the manufacturer's instructions. Pyroptotic cell death was assessed by performing double-staining with FLICA and propidium iodide (PI), followed by flow cytometry.
Patient Characteristics
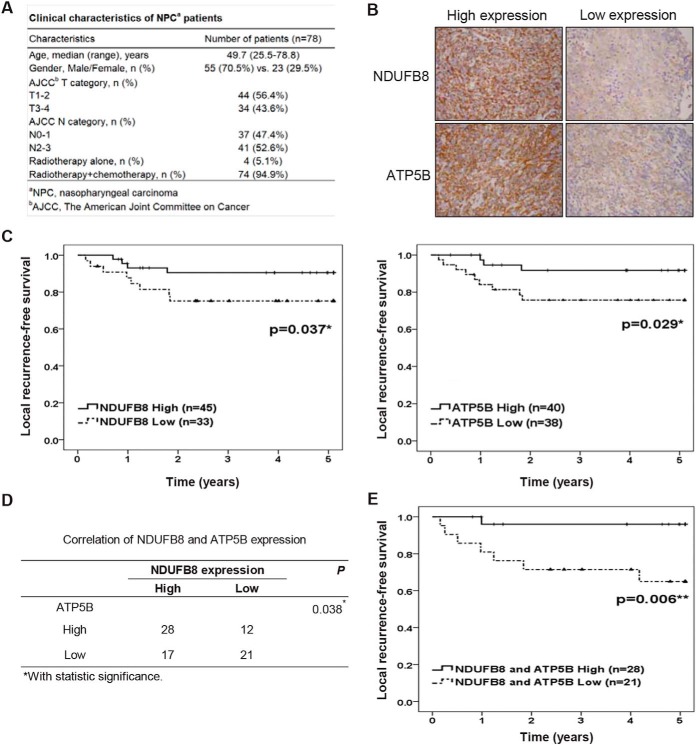
This study was conducted according to the principles expressed in the Declaration of Helsinki and was reviewed and approved by the Institutional Review Board (IRB) of Chang Gung Medical Foundation, Taoyuan, Taiwan. The specimens used for immunohistochemistry were harvested from 78 NPC patients seen at Chang Gung Memorial Hospital at Linkou, Taoyuan, Taiwan, from 2003 to 2013. Before sample collection, an IRB-approved informed consent form was signed by each participant. The median age at diagnosis was 49.7 years (range, 25.5–78.8), and the male-to-female ratio was ∼2.4:1. Clinical stage was defined according to the cancer staging system revised by the 7th edition of American Joint Committee on Cancer (AJCC) staging manual. All enrolled patients completed radiotherapy treatment. Among them, 74 patients had received additional chemotherapy. Any recurrence occurred at the primary site was a local recurrence. Local recurrence-free survival and overall survival was calculated as the time from the initiation of primary radiotherapy to the date of death or the last follow-up. Patient characteristics and clinical features are shown in Fig. 5A. The examination and enrolment of patients conducted as previously described (28).
Fig. 5.
Association of NDUFB8 and ATP5B with better survival in NPC patients. A, The clinical pathological parameters of the NPC patients. B, Expression levels of NDUFB8 and ATPT5B proteins in NPC tumor cells. Cells were immunohistochemically stained with protein-specific antibodies, and the results are demonstrated at 400x magnification. C, Kaplan-Meier survival analysis of local recurrence-free survival curves for NPC patients with NDUFB8 expression (left panel) or ATP5B expression (right panel). D, Correlation between NDUFB8 expression and ATP5B expression in NPC tissues. E, Kaplan-Meier survival analysis of local recurrence-free survival curves for NPC patients with combine expression of NDUFB8 and ATP5B. Symbols: *p < 0.05; **p < 0.01.
Immunohistochemistry (IHC)
IHC analysis was performed as previously described (6). We used primary antibodies against NDUFB8 (1:150; cat# HPA003886; Sigma-Aldrich) and ATP5B (1:100; cat# SC-55597; Santa Cruz Biotechnology). The staining intensity was classified as 0 (negative), 1 (weak), 2 (moderate) or 3 (strong). The H score with combining staining intensity and proportion of positive cells reflected protein expression and were used to classify the specimens/patients into two categories: “high-level” expression (NDUFB8 scores≥100 and/or ATP5B scores≥130) and “low-level” expression (NDUFB8 scores<100 and/or ATP5B scores<130). All cases were scored by an experienced pathologist.
Experimental Design and Statistical Rationale
For investigation of the NLRP3 inflammasome activation, NPC cells were treated with NLRP3-activating stimuli, including the cisplatin, nigericin, ATP, MSU or Alum. For iTRAQ-based LC-MS/MS analysis, ASC speck(−) and speck(+) NPC-HK1-ASC-GFP cells were analyzed in two independent biological replicates. The cutoff value for determining whether a protein was considered dysregulated was selected according to a comparison of protein levels between the same ASC speck(−) samples in two different batches. Proteins with a log2 ratio greater than the mean ratio plus one S.D. and those with a log2 ratio less than the mean ratio minus one S.D. were overexpressed and underexpressed, respectively. The functional enrichment analysis of identified proteins was predicted on GO terms analysis with the Metacore pathway analysis package or DAVID Bioinformatics Resources, which calculates enrichment p values in the different types of gene sets. We selected proteins whose iTRAQ ratios (ASC speck(+) cells versus ASC speck(−) cells) were at least 2SD above the mean in two biological duplicates, and subjected them to further verification using Western blotting. Additionally, NPC-BM1 cells, another NPC cell line were also used to investigate the difference in protein profiles between speck(+) and speck(−) cells through iTRAQ-based proteomic analysis.
Further, we further investigated whether the candidate proteins are essential for modulation of NLRP3 inflammasome activation and mitochondrial functions by gene knockdown with siRNA. We assessed their functions in NLRP3 inflammasome activation (NLRP3-mediated ASC speck formation, caspase-1 activation, IL-1β secretion, and cell pyroptosis) and mitochondrial activity (mtROS production) in our established cell model that the NLRP3 inflammasome is activated by cisplatin. Data are presented as the mean ± S.D. of three independent experiments and were analyzed with the Student's t test. p value less than 0.05 were considered as significant.
To determine the association between the expression of OxPhos proteins and clinical pathological parameters in NPC. The specimens harvested from 78 NPC patients were used for IHC analysis. The expression levels of target proteins were evaluated by assessing both the staining intensity and the percentage of staining-positive cells. Additionally, we looked for the correlation between the protein expression and patient survival. Statistical analysis was done using the PASW Statistics 18.0.0 statistical software package (SPSS Inc., 2009), as described previously (22). Survival analysis was performed using the Kaplan-Meier estimator. The log rank test was used to compare the survival curves of two groups and calculate the relevant p value. Multivariate analysis of survival using Cox proportional hazards models was conducted to identified which covariates were independently associated with local recurrence-free survival and overall survival. The relevant p value was calculated after cox regression. Further, the correlation between the expression levels of NDUFB8 and ATP5B was evaluated using Pearson's chi-squared test. A p value less than 0.05 was considered significant.
RESULTS
Comprehensive Proteomic Analysis of Differentially Expressed Proteins in NPC Cells with or Without ASC Specks
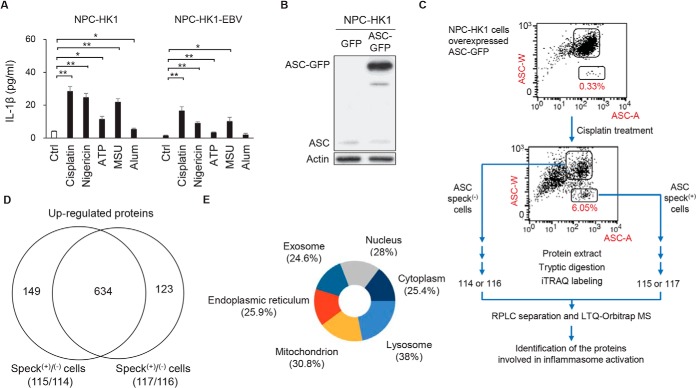
The inflammasome is a sensory complex that alters the immune system in response to pathogen- and damage-induced signals. To elucidate the activation of NLRP3 inflammasome after treatment with NLRP3-activating stimuli, we treated NPC-HK1 cells with NLRP3 stimuli, including the therapeutic drug cisplatin, pore-forming toxin nigericin, ATP, MSU crystal or Alum (6, 29–31). The secretion of IL-1β was significantly increased in NPC-HK1 cells treated with NLRP3 activators (left panel in Fig. 1A). Moreover, EBV infection has been reported to link with NPC, the EBV positive NPC-HK1-EBV cells were also tested for the inflammasome activation after treatment with NLRP3 activators (cisplatin, nigericin, ATP, MSU or Alum). Similar effects on IL-1β secretion were obtained when we used EBV-positive NPC cells (NPC-HK1-EBV) (right panel in Fig. 1A). The percentage of speck forming cells were increased in cells treated with cisplatin (from 0.25% to 5.59%), nigericin (from 0.35% to 3.05%), ATP (from 0.47% to 1.81%), MSU (from 0.19% to 0.70%) or Alum (from 0.16% to 0.31%) (supplemental Fig. S1).
Fig. 1.
iTRAQ analysis to identify proteins differentially expressed in NPC-HK1 cells with or without ASC specks. A, NPC-HK1 and NPC-HK1-EBV cells were treated with cisplatin (40 μm), nigericin (10 μm), ATP (5 mm), MSU (200 μm) or Alum (200 μm) for 24 h. The concentrations of IL-1β were measured in the cultured medium of cells by enzyme-linked immunosorbent assay (ELISA). Symbols: *p < 0.05; **p < 0.01. B, The ASC-GFP fusion protein was overexpressed in NPC-HK1 cells. Cell lysates were subjected to Western blotting for ASC protein and β-actin. C, Flow chart of the iTRAQ-based proteomics approach. NPC-HK-1-ASC-GFP cells were treated with cisplatin (40 μm) for 24 h, ASC speck-containing cells were isolated by flow cytometry (low and high pulse width (W) to pulse area (A) profile (ASC-W:ASC-A profile) were taken as indicating ASC speck(+) cells and ASC speck(−) cells, respectively), and iTRAQ- based quantitative proteomic analysis was performed. D, A total of 634 proteins were enriched (overexpressed) in speck(+) NPC-HK-1-ASC-GFP cells after cisplatin stimulation. E, Pie chart showing the cellular localization percentages of proteins found to be enriched (overexpressed) in speck(+) NPC-HK-1-ASC-GFP cells, as assessed using the Functional Enrichment Analysis tool (http://www.funrich.org/).
Previous studies showed that NLRP3 inflammasome function is determined by the homotypic domain interaction-mediated recruitment of proteins to the complex, which results in the formation of speck-like particles (32). However, the molecular mechanisms involved in NLRP3-ASC speck formation and inflammasome activation remain unclear. To address this gap, we herein established NPC-HK1 cells stably expressing an ASC-GFP fusion protein (hereafter referred to as NPC-HK1-ASC-GFP cells) (Fig. 1B). The NPC-HK1-ASC-GFP cells allowed us to visually monitor ASC speck formation in response to treatment with the various NLRP3 agonists (supplemental Fig. S1). Additionally, a significant increase of IL-1β secretion were demonstrated in NPC-HK1-ASC-GFP cells treated with NLRP3 activators (supplemental Fig. S1). Indeed, cells treated with cisplatin (∼6% speck(+) cells), which is often used to treat NPC exhibited higher percentage of ASC speck formation than other NLRP3 agonists, including nigericin (3.05%), ATP (1.81%), MSU (0.70%) or Alum (0.31%) (33) (lower panel in Fig. 1C and supplemental Fig. S1). To investigate the proteomic differences between the speck(+) and speck(−) cells, we isolated these cell populations by flow cytometry (25) and subjected them to iTRAQ-based proteomic analysis (Fig. 1C). The iTRAQ-based analysis resulted in 4820 and 4780 proteins which were identified (supplemental Table S1) and quantified (supplemental Table S2), respectively. A total of 634 proteins were found to be enriched (overexpressed) in speck(+) NPC-HK1-ASC-GFP cells after cisplatin stimulation (Fig. 1D and supplemental Table S3). Computational prediction revealed that more than 30% of the proteins enriched in the speck(+) cells were involved in mitochondrial functions (Fig. 1E and supplemental Table S4). These results indicated that mitochondrial proteins may play crucial roles in regulating NLRP3 inflammasome activation in NPC cells.
Mitochondrial OxPhos Components Are Enriched in ASC speck(+) NPC Cells
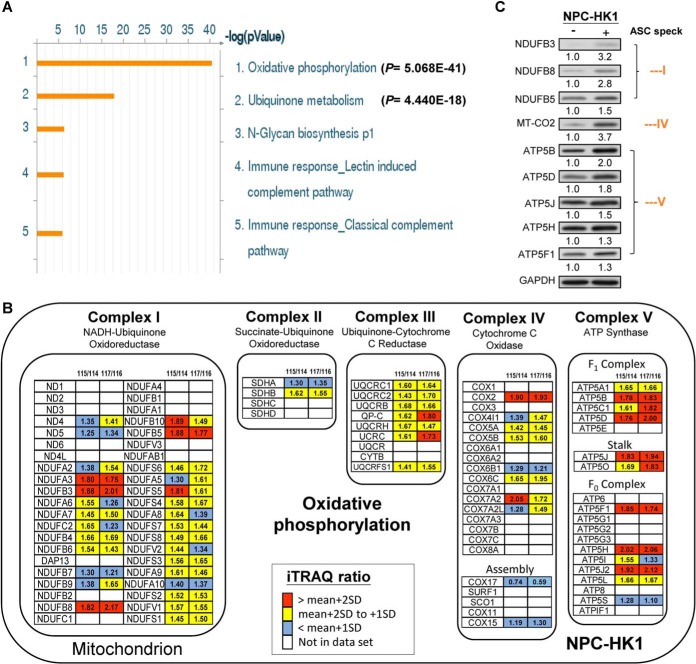
Gene ontology (GO) process network analysis of the 634 proteins found to be enriched in speck(+) versus speck(−) NPC cells revealed that most of them participated in two mitochondrion-based biological pathways: mitochondrial OxPhos (p = 5.068E-41) and ubiquinone metabolism (p = 4.440E-18) (Fig. 2A). We also observed that the levels of OxPhos proteins were higher in ASC speck(+) cells (Fig. 2B). Notably, the proteins found to be significantly enriched in ASC speck(+) cells were scattered among all five complexes of the OxPhos machinery, with higher proportions distributing to complexes I and V (Fig. 2B). We selected proteins whose iTRAQ ratios (ASC speck(+) cells versus ASC speck(−) cells) were at least 2SD above the mean in two biological duplicates, and subjected them to further verification using Western blotting. In ASC speck(+) cells, we detected increased levels of OxPhos-related subunits from complex I (NDUFB3, NDUFB8, and NDUFB5), complex IV (MT-CO2) and complex V (ATP5B, ATP5D, ATP5J, ATP5H, and ATP5F1) (Fig. 2C). Moreover, NPC-BM1 cells, another NPC cell line, which demonstrated increased IL-1β secretion and ASC speck formation in response to cisplatin stimulation (supplemental Fig. S2A–S2B) were also used to investigate the difference in protein profiles between speck(+) and speck(−) cells through iTRAQ-based proteomic analysis (supplemental Fig. S2B). The iTRAQ-based analysis resulted in 4803 and 4667 proteins which were identified (supplemental Table S5) and quantified (supplemental Table S6), respectively. A total of 559 proteins were found to be enriched (overexpressed) in speck(+) NPC-BM1-ASC-GFP cells after cisplatin stimulation (supplemental Table S7). Consistently, the results indicated that mitochondrial OxPhos components are enriched in ASC speck(+) NPC-BM1 cells (supplemental Fig. S2C–S2D and supplemental Table S8). These data strongly indicate that mitochondrial OxPhos components play a pivotal role in NLRP3 inflammasome formation and activation in NPC cells.
Fig. 2.
Mitochondrial OxPhos components are enriched in ASC speck(+) NPC cells. NPC-HK1-ASC-GFP cells were treated with cisplatin (40 μm) for 24 h and speck(+) and speck(−) cells were isolated using flow cytometry. A. GO process network of enriched proteins, as generated using the Metacore pathway analysis package. Most of the enriched or elevated proteins fell into two pathways: oxidative phosphorylation (p = 5.068E-41) and ubiquinone metabolism (p = 4.440E-18). B, List of OxPhos protein subunits identified by iTRAQ labeling and LC-MS/MS analysis. The iTRAQ ratios of 115/114 and 117/116 represent the quantitative ratio between ASC speck(+) (labeled with 115 or 117 iTRAQ reagents) and speck(−) (labeled with 114 or 116 iTRAQ reagents) cells. Red boxes indicate proteins with iTRAQ ratios of at least 2SD above the mean. Yellow boxes indicate proteins with iTRAQ ratios between 1 and 2 S.D. above the mean. Blue boxes indicate proteins that did not show a change in iTRAQ ratio. White boxes indicate proteins that were not identified in the proteome. Figure adapted from https://www.wikipathways.org/index.php/Pathway:WP111. C, Cell lysates from ASC speck(+) and ASC speck(−) NPC-HK1-ASC-GFP cells were used for Western blot analysis. The ratio of OxPhos proteins were calculated by normalization with GAPDH, with the values from the ASC speck(−) cells set as 1.0.
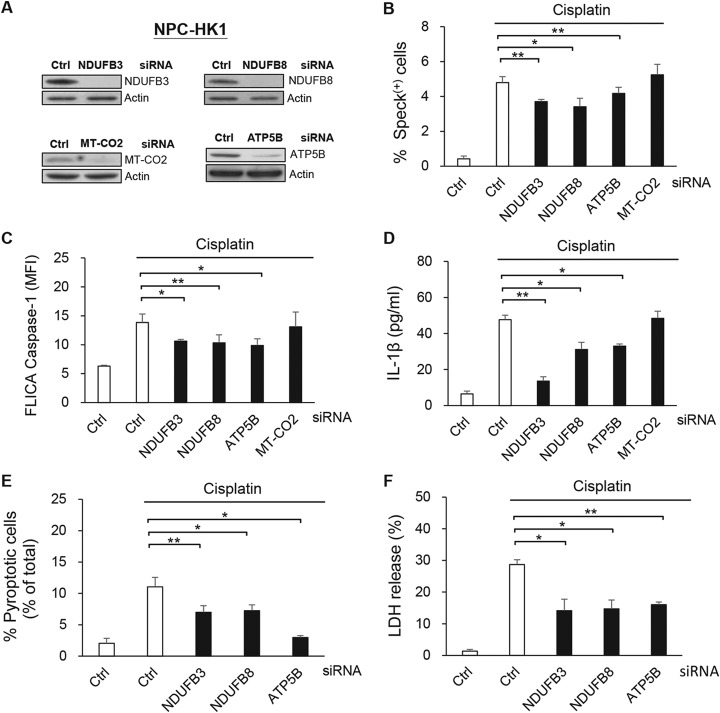
NDUFB3, NDUFB8, and ATP5B are Involved in NLRP3 Inflammasome Activation and Cell Pyroptosis
Four proteins whose levels were >2 fold enriched in speck(+) cells (NDUFB3, NDUFB8, ATP5B and MT-CO2) were further verified by treating NPC-HK1-ASC-GFP cells with gene-specific siRNAs (Fig. 3A), and then further treating them with cisplatin. As presented in Fig. 3B, the percentage of ASC-speck(+) cells was significantly reduced in cells treated with siRNAs against NDUFB3, NDUFB8 and ATP5B, but not MT-CO2 (Fig. 3B). We used the FLICA reagent, which forms a covalent bond with the active protease (34), to monitor the activation of caspase-1, which is a step in the process of inflammasome activation. As demonstrated in Fig. 3C, the cisplatin-induced activity of caspase-1 was decreased in NPC-HK1 cells subjected to knockdown of NDUFB3, NDUFB8 or ATP5B, compared with cells treated with the control siRNA. Moreover, IL-1β secretion was significantly decreased in NDUFB3-, NDUFB8- or ATP5B-knockdown NPC-HK1 cells compared with control cells (Fig. 3D). Consistently, similar results were obtained in NPC-BM1 cells (supplemental Fig. S3A–S3D). In contrast, siRNAs directed against less strongly enriched proteins (i.e. NDUFB5, ATP5H or ATP5J) (supplemental Fig. S4A) did not significantly alter cisplatin-induced ASC speck formation (supplemental Fig. S4B), caspase-1 activation (supplemental Fig. S4C) or IL-1β secretion (supplemental Fig. S4D). Collectively, these results indicate that mitochondrial NDUFB3, NDUFB8, and ATP5B participate in modulating NLRP3 inflammasome activation.
Fig. 3.
NDUFB3, NDUFB8 and ATP5B are involved in NLRP3 inflammasome activation and cell pyroptosis. NPC-HK1 cells were transfected with NDUFB3-, NDUFB8-, ATP5B-, MT-CO2, or control (Ctrl)- siRNA for 48 h, treated with cisplatin (40 μm) for 24 h and analyzed for ASC speck formation, caspase-1 activity, IL-1β release and cell pyroptosis. A, Lysates were collected from knockdown cells and used for Western blot analysis. B, The percentages of ASC speck(+) cells were quantified from NPC-HK1-ASC-GFP cells by flow cytometry. C, Activated caspase-1 was determined by staining with YVAD-FLICA followed by flow cytometry. D, The levels of IL-1β in cell culture supernatants were measured by ELISA. E, The percentages of pyroptotic cells (FLICA-positive/PI-positive cells) were detected by flow cytometry. F, Supernatant LDH activity was assessed using an LDH Cytotoxicity Assay Kit and the percentages of LDH release are presented in a histogram. Symbols: *p < 0.05; **p < 0.01.
To assess pyroptotic cell death, which is a form of lytic programmed cell death that is triggered by inflammasome activation, we double stained cells for activated caspase-1 and nuclei (using PI) or assessed their release of LDH (35). As shown in Fig. 3E, the proportion of activated caspase-1/PI double-positive NPC-HK1 cells was significantly lower in NDUFB3-, NDUFB8-, or ATP5B-knockdown cells compared with control cells following cisplatin-induced inflammasome activation. Similarly, these knockdown cells exhibited decreased LDH release relative to the siRNA control group (Fig. 3F). Similar results were demonstrated in NPC-BM1 cells (supplemental Fig. S3E–S3F). Taken together, our results reveal that the mitochondrial OxPhos proteins, NDUFB3, NDUFB8 and ATP5B, appear to be involved in inflammasome activation and play important roles in regulating inflammasome-related cell pyroptosis.
NDUFB3, NDUFB8, and ATP5B Activate the NLRP3 Inflammasome by Regulating mtROS Production
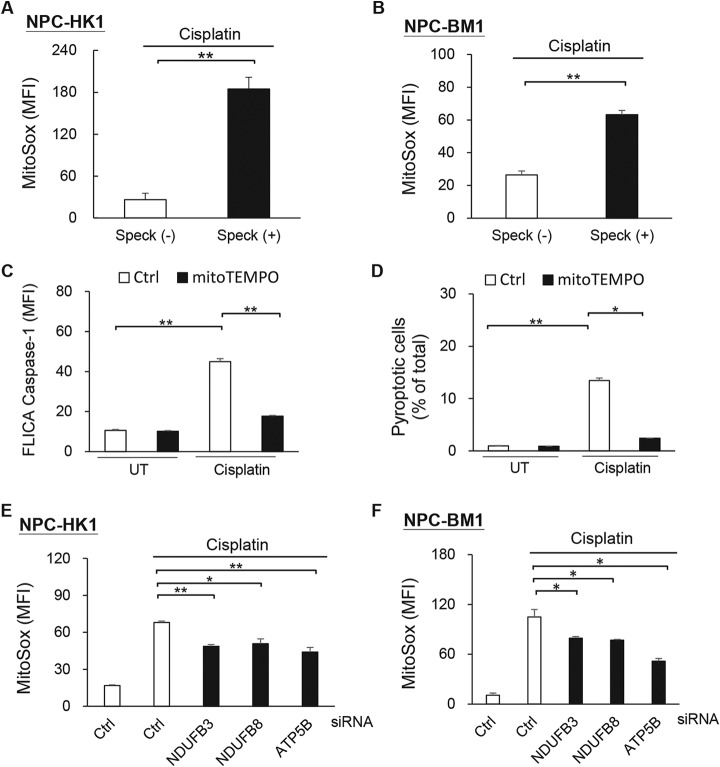
Although the above-described results strongly suggested that the mitochondrial OxPhos proteins are involved in regulating NLRP3 inflammasome activation, the mechanism(s) linking mitochondria to NLRP3 inflammasome activation in NPC cells remained unclear. Studies have shown that the NLRP3 inflammasome is activated by mtROS (36, 37), the mitochondrial OxPhos machinery is the major site of mtROS production, and mitochondrial dysfunctions leads to mtROS elevation (38). Therefore, we examined the effects of NDUFB3, NDUFB8, and ATP5B on the mtROS production triggered by NLRP3 inflammasome activation. Our experiments revealed that the levels of mtROS were elevated in ASC speck(+) NPC-HK1 and NPC-BM1 cells as compared with ASC speck(−) group (Fig. 4A–4B), and that caspase-1 activation (Fig. 4C) and pyroptotic cell death (Fig. 4D) were significantly inhibited by pretreatment with the mtROS scavenger, MitoTEMPO. Moreover, NDUFB3-, NDUFB8- and ATP5B-knockdown NPC cells exhibited significant reductions in cisplatin-induced mtROS production (Fig. 4E–4F). Together, these data suggest that mtROS is an upstream regulator of NLRP3 inflammasome activation in NPC cells, and that mitochondrial OxPhos proteins may mediate mtROS production in these cells.
Fig. 4.
NDUFB3, NDUFB8 and ATP5B activate the NLRP3 inflammasome by regulating mtROS production. A, Production of mtROS by cisplatin-treated NPC-HK1-ASC-GFP cells with or without ASC specks. B, Production of mtROS by cisplatin-treated NPC-BM1-ASC-GFP cells with or without ASC specks. NPC-HK1 cells were pretreated with MitoTEMPO for 1 h and treated with cisplatin (40 μm) for 24 h. C, Activated caspase-1 was determined by staining with YVAD-FLICA followed by flow cytometry; D, The percentages of pyroptotic (FLICA-positive/PI-positive) cells were detected by flow cytometry. NPC-HK1 (E) and NBC-BM1 cells (F) were transfected with NDUFB3-, NDUFB8-, ATP5B-, or control (Ctrl)- siRNA for 48 h, treated with cisplatin (40 μm) for 24 h (NPC-HK1) or 30 h (NPC-BM1), and subjected to analysis of mtROS production. Symbols: *p < 0.05; **p < 0.01.
Association of NDUFB8 and ATP5B with Better Survival in NPC Patients
We previously reported that higher levels of inflammasome protein components (e.g. NLRP3) are significantly correlated with better post-treatment recurrence-free survival of NPC patients (6). In the present study, we show that OxPhos proteins are involved in modulating NLRP3 inflammasome activation and pyroptotic cell death in NPC cells. To examine whether these inflammasome activation-related OxPhos proteins are associated with clinical pathological parameters, we examined 78 biopsy specimens from NPC patients (for a summary of clinical characteristics, see Fig. 5A). We performed IHC analysis to assess the expression levels of NDUFB8 and ATP5B in tumors. We then used the expression levels of NDUFB8 (upper two panels in Fig. 5B) and ATP5B (lower two panels in Fig. 5B) to classify the NPC tumor samples into high- or low-expression groups and analyzed the association of each group with patient survival. Kaplan-Meier survival analyses revealed that better local recurrence-free survival was seen among NPC patients in the high-expression groups for NDUFB8 (p = 0.037; left panel in Fig. 5C) and ATP5B (p = 0.029; right panel in Fig. 5C). Because our results indicated that NDUFB8 and ATP5B can modulate NLRP3 inflammasome activation and cell pyroptosis, we investigated whether their expression levels were correlated, and whether there was prognostic value in assessing the combined expression of these proteins. Indeed, we found that the expression of NDUFB8 and was positively correlated with that of ATP5B in NPC tissues (Fig. 5D), and that high-level expression of both NDUFB8 and ATP5B in the tumor was strongly associated with better local recurrence-free survival of patients (p = 0.006, Fig. 5E). We next conducted a multivariate analysis of NDUFB8 and ATP5B expression with clinicopathological features, including tumor (T) stage and node (N) stage, and found that high-ATP5B expression (p = 0.047) was a marker of better local recurrence-free survival (Table I). Although high-NDUFB8 expression was not an independent predictor of better local recurrence-free survival in multivariate analysis (p > 0.05), combined high expression of NDUFB8 and ATP5B served as marker for better local recurrence-free survival (p = 0.035). However, Kaplan-Meier survival analyses (supplemental Fig. S5A–S5C) and multivariate analysis (supplemental Fig. S5D) showed no significant association between the expression of NDUFB8 and ATP5B with overall survival of NPC patients. Together, the results of our clinical pathological analysis indicate that NDUFB8 and ATP5B could potentially be developed as useful biomarkers for local recurrence in NPC.
Table I. Multivariate analysis of the association between NDUFB8 and ATP5B expression with local recurrence-free survival of NPC patients.
| Characteristics | Hazards Ratio (95% CI) | p value |
|---|---|---|
| Patients (n = 49) | ||
| T stage (3–4 vs. 1–2) | 0.543 (0.125–2.367) | 0.416 |
| N stage (2–3 vs. 0–1) | 1.012 (0.230–4.453) | 0.987 |
| NDUFB8+ATP5B (High vs. Low) | 0.102 (0.012–0.848) | 0.035 |
| Patients (n = 78) | ||
| T stage (3–4 vs. 1–2) | 1.126 (0.377–3.359) | 0.831 |
| N stage (2–3 vs. 0–1) | 1.377 (0.444–4.273) | 0.580 |
| NDUFB8 (High vs. Low) | 0.324 (0.098–1.070) | 0.065 |
| Patients (n = 78) | ||
| T stage (3–4 vs. 1–2) | 1.169 (0.390–3.503) | 0.780 |
| N stage (2–3 vs. 0–1) | 1.511 (0.489–4.663) | 0.473 |
| ATP5B (High vs. Low) | 0.269 (0.074–0.980) | 0.047 |
The Cox proportional hazards model was applied for multivariate analysis to determine the independence of each prognostic factor.
DISCUSSION
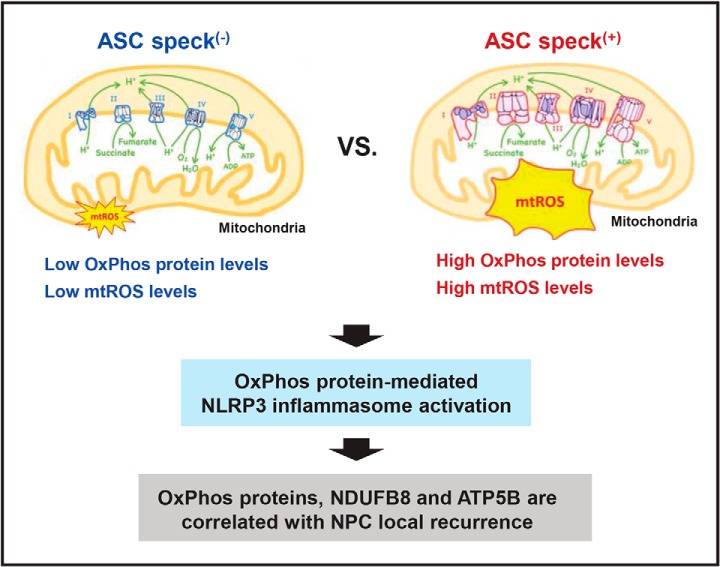
In this report, we demonstrate that mitochondria are linked to NLRP3 inflammasome activation through mitochondrial OxPhos proteins. Using flow cytometry analysis and iTRAQ technology, we isolated ASC speck(+) cells and found that they are enriched with OxPhos proteins, which modulate mitochondrial mtROS production and downstream inflammasome activation. We further found that NDUFB8 (complex I) and ATP5B (complex V) serve as markers for local recurrence-free survival in NPC patients. This study is the first to experimentally link mitochondrial OxPhos proteins, inflammasome activation and clinical outcomes among NPC patients (Fig. 6).
Fig. 6.
Role of mitochondrial OxPhos proteins in NLRP3 inflammasome activation in NPC. Based on the results of our comprehensive proteomic and functional studies, we propose that mitochondrial OxPhos components are enriched in ASC speck-forming NPC cells, enhance mtROS levels and thereby contribute to NLRP3 inflammasome formation and activation. High expression levels of OxPhos components, including NDUFB8 (complex I) and ATP5B (complex V), are correlated with better local recurrence-free survival in NPC patients and thus might prove useful as markers for NPC local recurrence.
ASC speck formation, which is a hallmark for inflammasome activation, can be controlled by various cellular events, including the phosphorylation or ubiquitination of ASC molecules (39, 40). We previously demonstrated that Syk kinase can phosphorylate Pyk2, the phosphorylated Pyk2 relocalizes to ASC specks upon NLRP3 inflammasome activation, Pyk2 can directly phosphorylate ASC at Tyr146, and only phosphorylated ASC can participate in speck formation and trigger IL-1β secretion (27). Other reports have shown that linear ubiquitination assembly complex and TNFR-associated factor 3 are required for ASC-ubiquitination-mediated assembly of the NLRP3/ASC inflammasome (41, 42). However, we do not yet fully understand the signaling networks that contribute to assembling and regulating inflammasome speck formation. Here, we comprehensively compared the protein expression patterns of ASC speck(+) and speck(−) NPC cells, and found that mitochondrial OxPhos proteins are critical for inflammasome formation and activation in this system. In addition, we found that the ASC speck(+) NPC cells were enriched for various mitochondrial proteins, including some with functions in cristae formation, fatty acid β-oxidation and mitochondrial protein import. These data strongly suggest that there is a link between mitochondrial functions and NLRP3 inflammasome formation/activation.
Mitochondrial dysfunction is a major event in NLRP3 inflammasome activation, as NLRP3 stimuli trigger the mitochondrial OxPhos machinery to increase mtROS and decrease mtΔΨ (13, 15). It is known that OxPhos complexes I, III and IV form a respiratory supercomplex (43) that plays a key role in regulating mtROS production (44). OxPhos complex V, which is a major site for intracellular ATP generation, may also increase mtROS production; conversely the activity of OxPhos complex V can be disrupted by the specific ATP synthase inhibitor, oligomycin (45). In the present study, we observed higher levels of OxPhos proteins and mtROS production in ASC speck(+) NPC cells, suggesting that the former two parameters are correlated with inflammasome activity. Our findings are consistent with the previous report that ovarian cancer cell lines with high levels of OxPhos proteins (high-OxPhos) displayed higher levels of ROS and enhanced chemosensitivity to platinum salts and taxane, compared with low-OxPhos cells (46). Thus, the accumulation of high levels of ROS in high-OxPhos cells is likely to be a key determinant in activating the NLRP3 inflammasome and thereby increasing the chemosensitivity of cancer cells.
We previously demonstrated that high-level NLRP3 inflammasome expression is correlated with better post-treatment survival in NPC patients (6), but the mechanism underlying this effect was unclear. ROS have been positively linked to NLRP3 inflammasome activation and the initiation, promotion, progression and metastasis of cancer (47). Various cancer chemotherapeutics (e.g. cisplatin) alter the mitochondrial-dependent ROS response to trigger cell death by greatly enhancing the mtROS level (48). In the present study, our systematic analysis of OxPhos complex components revealed that in cisplatin-stimulated NPC cells, NDUFB3, NDUFB8, and ATP5B modulate mtROS production, leading to NLRP3 inflammasome activation and cell pyroptosis. Importantly, we observed an apparent correlation between high expression levels of NDUFB8 and ATP5B in NPC tumor specimens, and this positive relationship seemed to be correlated with better survival outcomes in NPC patients. These findings suggest that NDUFB8 and ATP5B may cooperate in facilitating the NLRP3 inflammasome-mediated regulation of NPC treatment. Consistent with our findings, higher-level expression of ATP5B has been correlated with better survival in gallbladder cancer (49) and acute myeloid leukemia (50). Therefore, the previous findings and our present results indicate that NLRP3, ATPT5B, and NDUFB8 can be regarded as markers for assessment of local recurrence in NPC patients.
In summary, we herein describe a previously unrecognized mitochondrial OxPhos component/mtROS axis and show that it modulates NLRP3 inflammasome formation and activation. We further propose that high-level expression of inflammasome-related OxPhos proteins could be developed as biomarkers for local recurrence in NPC patients and/or targeted as a new therapeutic strategy against inflammation-associated cancer.
DATA AVAILABILITY
The MS raw data for iTRAQ-based proteome analysis were deposited on the ProteomeXchange Consortium website (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (51), data set identifier: PXD013071. The information of identified proteins are provided as the supplemental data.
Supplementary Material
Footnotes
* This work was supported by grants to Y.S.C. from the Ministry of Science and Technology (MOST), Taiwan (MOST 105–2320-B-182–034-MY3, 105–2811-B-182–029, 106–2811-B-182–015, and 107–2811-B-182–521) and Chang Gung Memorial Hospital (CGMH), Taiwan (CMRPD1D0101–103 and BMRP021), to L.C.C. from MOST, Taiwan (MOST 105–2628-B-715–002-MY3), Mackay Medical College, Taiwan (1061B25 and 1071B26), and the Mackay Memorial Hospital, Taiwan (MMH-MM-10702), and to C.C.W. from MOST, Taiwan (MOST 107–2320-B-182–009) and CGMH, Taiwan (CLRPD190018 and BMRPC77), by grants to the Ministry of Education, Taiwan to Chang Gung University (EMRPD1G0031). The work was also supported by grants from Research Center for Emerging Viral Infections, Chang Gung University, and Molecular Medicine Research Center, Chang Gung University, from Featured Areas Research Center Program within the Framework of Higher Education Sprout Project by Ministry of Education and Ministry of Science and Technology, Taiwan (MOST 107–3017-F-182–001). The authors declare that they have no conflicts of interest with the contents of this article.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ASC
- apoptosis-associated speck-like protein containing a caspase-recruitment domain
- CID
- collision-induced dissociations
- EB1
- end-binding protein 1
- EBV
- Epstein-Barr virus
- ETC
- electron transport chain
- FLICA
- fluorochrome labeled inhibitors of caspase-1
- GO
- Gene ontology
- HCD
- higher-energy collision-induced dissociations
- IHC
- immunohistochemistry
- IRB
- Institutional Review Board
- LDH
- lactate dehydrogenase
- LMP1
- latent membrane protein 1
- MAVS
- mitochondrial antiviral signaling protein
- Mfn2
- mitofusin 2
- mtDNA
- mitochondrial DNA
- mtΔΨ
- mitochondrial membrane potential
- mtROS
- mitochondrial reactive oxygen species
- NPC
- nasopharyngeal carcinoma
- OxPhos
- oxidative phosphorylation
- PI
- propidium iodide
- PYCARD
- PYD and CARD domain containing
- SD
- standard deviation
- TEABC
- triethylammonium bicarbonate
- TFA
- trifluoroacetic acid.
REFERENCES
- 1. He Q., Fu Y., Tian D., and Yan W. (2018) The contrasting roles of inflammasomes in cancer. Am. J. Cancer Res. 8, 566–583 [PMC free article] [PubMed] [Google Scholar]
- 2. Moossavi M., Parsamanesh N., Bahrami A., Atkin S. L., and Sahebkar A. (2018) Role of the NLRP3 inflammasome in cancer. Mol. Cancer 17, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Awad F., Assrawi E., Louvrier C., Jumeau C., Georgin-Lavialle S., Grateau G., Amselem S., Giurgea I., and Karabina S. A. (2018) Inflammasome biology, molecular pathology and therapeutic implications. Pharmacol. Therapeutics 187, 133–149 [DOI] [PubMed] [Google Scholar]
- 4. Hoss F., Rodriguez-Alcazar J. F., and Latz E. (2017) Assembly and regulation of ASC specks. Cell. Mol. Life Sci. 74, 1211–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kovacs S. B., and Miao E. A. (2017) Gasdermins: effectors of pyroptosis. Trends Cell Biol. 27, 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen L. C., Wang L. J., Tsang N. M., Ojcius D. M., Chen C. C., Ouyang C. N., Hsueh C., Liang Y., Chang K. P., Chen C. C., and Chang Y. S. (2012) Tumour inflammasome-derived IL-1beta recruits neutrophils and improves local recurrence-free survival in EBV-induced nasopharyngeal carcinoma. EMBO Mol. Med. 4, 1276–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang Y. T., Sheen T. S., Chen C. L., Lu J., Chang Y., Chen J. Y., and Tsai C. H. (1999) Profile of cytokine expression in nasopharyngeal carcinomas: a distinct expression of interleukin 1 in tumor and CD4+ T cells. Cancer Res. 59, 1599–1605 [PubMed] [Google Scholar]
- 8. Wang L. J., Hsu C. W., Chen C. C., Liang Y., Chen L. C., Ojcius D. M., Tsang N. M., Hsueh C., Wu C. C., and Chang Y. S. (2012) Interactome-wide analysis identifies end-binding protein 1 as a crucial component for the speck-like particle formation of activated absence in melanoma 2 (AIM2) inflammasomes. Mol. Cell. Proteomics 11, 1230–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang L. J., Huang H. Y., Huang M. P., Liou W., Chang Y. T., Wu C. C., Ojcius D. M., and Chang Y. S. (2014) The microtubule-associated protein EB1 links AIM2 inflammasomes with autophagy-dependent secretion. J. Biol. Chem. 289, 29322–29333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cai T. T., Ye S. B., Liu Y. N., He J., Chen Q. Y., Mai H. Q., Zhang C. X., Cui J., Zhang X. S., Busson P., Zeng Y. X., and Li J. (2017) LMP1-mediated glycolysis induces myeloid-derived suppressor cell expansion in nasopharyngeal carcinoma. PLoS Pathogens 13, e1006503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weinberg S. E., Sena L. A., and Chandel N. S. (2015) Mitochondria in the regulation of innate and adaptive immunity. Immunity 42, 406–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Q., Zhang D., Hu D., Zhou X., and Zhou Y. (2018) The role of mitochondria in NLRP3 inflammasome activation. Mol. Immunol. 103, 115–124 [DOI] [PubMed] [Google Scholar]
- 13. Zhong Z., Umemura A., Sanchez-Lopez E., Liang S., Shalapour S., Wong J., He F., Boassa D., Perkins G., Ali S. R., McGeough M. D., Ellisman M. H., Seki E., Gustafsson A. B., Hoffman H. M., Diaz-Meco M. T., Moscat J., and Karin M. (2016) NF-kappaB restricts inflammasome activation via elimination of damaged mitochondria. Cell 164, 896–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Horng T. (2014) Calcium signaling and mitochondrial destabilization in the triggering of the NLRP3 inflammasome. Trends Immunol. 35, 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou R., Yazdi A. S., Menu P., and Tschopp J. (2011) A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 [DOI] [PubMed] [Google Scholar]
- 16. Zhong Z., Liang S., Sanchez-Lopez E., He F., Shalapour S., Lin X. J., Wong J., Ding S., Seki E., Schnabl B., Hevener A. L., Greenberg H. B., Kisseleva T., and Karin M. (2018) New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature 560, 198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iyer S. S., He Q., Janczy J. R., Elliott E. I., Zhong Z., Olivier A. K., Sadler J. J., Knepper-Adrian V., Han R., Qiao L., Eisenbarth S. C., Nauseef W. M., Cassel S. L., and Sutterwala F. S. (2013) Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity 39, 311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Subramanian N., Natarajan K., Clatworthy M. R., Wang Z., and Germain R. N. (2013) The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell 153, 348–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ichinohe T., Yamazaki T., Koshiba T., and Yanagi Y. (2013) Mitochondrial protein mitofusin 2 is required for NLRP3 inflammasome activation after RNA virus infection. Proc. Natl. Acad. Sci. U.S.A. 110, 17963–17968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Triantafilou K., Hughes T. R., Triantafilou M., and Morgan B. P. (2013) The complement membrane attack complex triggers intracellular Ca2+ fluxes leading to NLRP3 inflammasome activation. J. Cell Sci. 126, 2903–2913 [DOI] [PubMed] [Google Scholar]
- 21. Mills E. L., Kelly B., and O'Neill L. A. J. (2017) Mitochondria are the powerhouses of immunity. Nature Immunol. 18, 488–498 [DOI] [PubMed] [Google Scholar]
- 22. Chung I. C., Yuan S. N., OuYang C. N., Lin H. C., Huang K. Y., Chen Y. J., Chung A. K., Chu C. L., Ojcius D. M., Chang Y. S., and Chen L. C. (2018) Src-family kinase-Cbl axis negatively regulates NLRP3 inflammasome activation. Cell Death Dis. 9, 1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lo A. K., Lo K. W., Tsao S. W., Wong H. L., Hui J. W., To K. F., Hayward D. S., Chui Y. L., Lau Y. L., Takada K., and Huang D. P. (2006) Epstein-Barr virus infection alters cellular signal cascades in human nasopharyngeal epithelial cells. Neoplasia 8, 173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liao S. K., Perng Y. P., Shen Y. C., Chung P. J., Chang Y. S., and Wang C. H. (1998) Chromosomal abnormalities of a new nasopharyngeal carcinoma cell line (NPC-BM1) derived from a bone marrow metastatic lesion. Cancer Genet. Cytogenet. 103, 52–58 [DOI] [PubMed] [Google Scholar]
- 25. Sester D. P., Thygesen S. J., Sagulenko V., Vajjhala P. R., Cridland J. A., Vitak N., Chen K. W., Osborne G. W., Schroder K., and Stacey K. J. (2015) A novel flow cytometric method to assess inflammasome formation. J. Immunol. 194, 455–462 [DOI] [PubMed] [Google Scholar]
- 26. Lin M. H., Li C. C., Shu J. C., Chu H. W., Liu C. C., and Wu C. C. (2018) Exoproteome profiling reveals the involvement of the foldase PrsA in the cell surface properties and pathogenesis of Staphylococcus aureus. Proteomics 18, e1700195. [DOI] [PubMed] [Google Scholar]
- 27. Chung I. C., OuYang C. N., Yuan S. N., Li H. P., Chen J. T., Shieh H. R., Chen Y. J., Ojcius D. M., Chu C. L., Yu J. S., Chang Y. S., and Chen L. C. (2016) Pyk2 activates the NLRP3 inflammasome by directly phosphorylating ASC and contributes to inflammasome-dependent peritonitis. Sci. Rep. 6, 36214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hsu C. W., Chen Y. T., Hsieh Y. J., Chang K. P., Hsueh P. C., Chen T. W., Yu J. S., Chang Y. S., Li L., and Wu C. C. (2019) Integrated analyses utilizing metabolomics and transcriptomics reveal perturbation of the polyamine pathway in oral cavity squamous cell carcinoma. Anal. Chim. Acta 1050, 113–122 [DOI] [PubMed] [Google Scholar]
- 29. Martinon F., Petrilli V., Mayor A., Tardivel A., and Tschopp J. (2006) Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440, 237–241 [DOI] [PubMed] [Google Scholar]
- 30. Hornung V., Bauernfeind F., Halle A., Samstad E. O., Kono H., Rock K. L., Fitzgerald K. A., and Latz E. (2008) Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nature Immunol. 9, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Katsnelson M. A., Rucker L. G., Russo H. M., and Dubyak G. R. (2015) K+ efflux agonists induce NLRP3 inflammasome activation independently of Ca2+ signaling. J. Immunol. 194, 3937–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lu A., Magupalli V. G., Ruan J., Yin Q., Atianand M. K., Vos M. R., Schroder G. F., Fitzgerald K. A., Wu H., and Egelman E. H. (2014) Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156, 1193–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sze H., Blanchard P., Ng W. T., Pignon J. P., and Lee A. W. (2015) Chemotherapy for nasopharyngeal carcinoma - current recommendation and controversies. Hematol. Oncol. Clin. North Am. 29, 1107–1122 [DOI] [PubMed] [Google Scholar]
- 34. Grabarek J., and Darzynkiewicz Z. (2002) In situ activation of caspases and serine proteases during apoptosis detected by affinity labeling their enzyme active centers with fluorochrome-tagged inhibitors. Exp. Hematol. 30, 982–989 [DOI] [PubMed] [Google Scholar]
- 35. Rayamajhi M., Zhang Y., and Miao E. A. (2013) Detection of pyroptosis by measuring released lactate dehydrogenase activity. Methods Mol. Biol. 1040, 85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Abais J. M., Xia M., Zhang Y., Boini K. M., and Li P. L. (2015) Redox regulation of NLRP3 inflammasomes: ROS as trigger or effector? Antioxidants Redox Signal. 22, 1111–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sandhir R., Halder A., and Sunkaria A. (2017) Mitochondria as a centrally positioned hub in the innate immune response. Biochim. Biophys. Acta 1863, 1090–1097 [DOI] [PubMed] [Google Scholar]
- 38. Zorov D. B., Juhaszova M., and Sollott S. J. (2014) Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 94, 909–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kattah M. G., Malynn B. A., and Ma A. (2017) Ubiquitin-modifying enzymes and regulation of the inflammasome. J. Mol. Biol. 429, 3471–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Song N., and Li T. (2018) Regulation of NLRP3 inflammasome by phosphorylation. Front. Immunol. 9, 2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rodgers M. A., Bowman J. W., Fujita H., Orazio N., Shi M., Liang Q., Amatya R., Kelly T. J., Iwai K., Ting J., and Jung J. U. (2014) The linear ubiquitin assembly complex (LUBAC) is essential for NLRP3 inflammasome activation. J. Exp. Med. 211, 1333–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guan K., Wei C., Zheng Z., Song T., Wu F., Zhang Y., Cao Y., Ma S., Chen W., Xu Q., Xia W., Gu J., He X., and Zhong H. (2015) MAVS Promotes Inflammasome Activation by Targeting ASC for K63-Linked Ubiquitination via the E3 Ligase TRAF3. J.Immunol. 194, 4880–4890 [DOI] [PubMed] [Google Scholar]
- 43. Chaban Y., Boekema E. J., and Dudkina N. V. (2014) Structures of mitochondrial oxidative phosphorylation supercomplexes and mechanisms for their stabilisation. Biochim. Biophys. Acta 1837, 418–426 [DOI] [PubMed] [Google Scholar]
- 44. Enriquez A., R-P. (2014) The function of the respiratory supercomplexes: The plasticity model. Biochim. Biophys. Acta 1837, 444–450 [DOI] [PubMed] [Google Scholar]
- 45. Vaamonde-Garcia C., Loureiro J., Valcarcel-Ares M. N., Riveiro-Naveira R. R., Ramil-Gomez O., Hermida-Carballo L., Centeno A., Meijide-Failde R., Blanco F. J., and Lopez-Armada M. J. (2017) The mitochondrial inhibitor oligomycin induces an inflammatory response in the rat knee joint. BMC Musculoskelet. Disord. 18, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gentric G., Kieffer Y., Mieulet V., Goundiam O., Bonneau C., Nemati F., Hurbain I., Raposo G., Popova T., Stern M. H., Lallemand-Breitenbach V., Muller S., Caneque T., Rodriguez R., Vincent-Salomon A., de The H., Rossignol R., and Mechta-Grigoriou F. (2019) PML-regulated mitochondrial metabolism enhances chemosensitivity in human ovarian cancers. Cell Metab. 29, 156–173 e110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang H., Villani R. M., Wang H., Simpson M. J., Roberts M. S., Tang M., and Liang X. (2018) The role of cellular reactive oxygen species in cancer chemotherapy. J. Exp. Clin. Cancer Res. 37, 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marullo R., Werner E., Degtyareva N., Moore B., Altavilla G., Ramalingam S. S., and Doetsch P. W. (2013) Cisplatin induces a mitochondrial-ROS response that contributes to cytotoxicity depending on mitochondrial redox status and bioenergetic functions. PloS One 8, e81162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sun J., Yang Z. L., Miao X., Zou Q., Li J., Liang L., Zeng G., and Chen S. (2015) ATP5b and beta2-microglobulin are predictive markers for the prognosis of patients with gallbladder cancer. J. Mol. Histol. 46, 57–65 [DOI] [PubMed] [Google Scholar]
- 50. Xiao X., Yang J., Li R., Liu S., Xu Y., Zheng W., Yi Y., Luo Y., Gong F., Peng H., Pei M., Deng M., and Zhang G. (2013) Deregulation of mitochondrial ATPsyn-beta in acute myeloid leukemia cells and with increased drug resistance. PloS One 8, e83610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vizcaino J. A., Csordas A., Del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, 11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS raw data for iTRAQ-based proteome analysis were deposited on the ProteomeXchange Consortium website (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (51), data set identifier: PXD013071. The information of identified proteins are provided as the supplemental data.