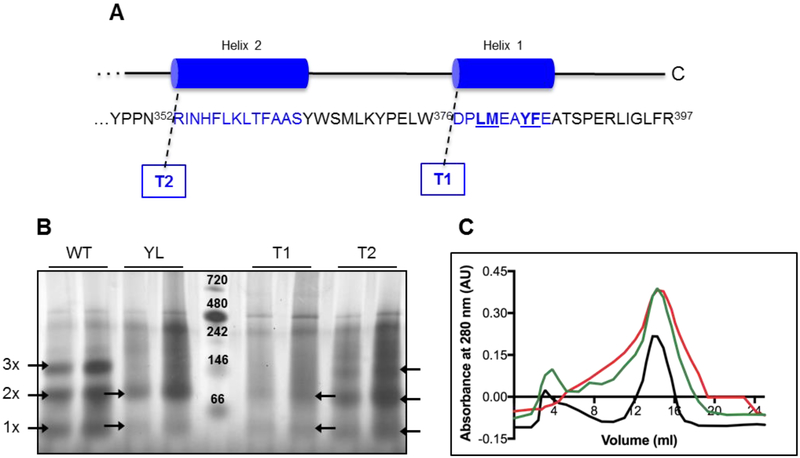
Figure 1. Oligomeric state of WT and soluble mutants by Blue Native-PAGE.
(A) Soluble mutants were previously constructed by truncation of one (T1) or both (T2) C-terminal helices, and by point mutations of the four amino acids underlined in the last helix (YL) [12]. (B) 5 (left) and 10 μg (right) of each protein sample were loaded in native buffer plus Coomassie Brilliant Blue G. Arrows indicate bands corresponding to the monomer (1x), dimer (2x) and trimer (3x) complexes (about 46, 88 and 139 kDa, respectively). The bands of the protein ladder (center lane) represent: Apoferritin band 1 (720 kDa), Apoferritin band 2 (480 kDa), β-phycoerythrin (242 kDa), lactate dehydrogenase (146 kDa), bovine serum albumin (66 kDa); from top to bottom. (C) Chromatograms of the purified WT (black), T1 (green) and T2 (red) SQRs showing the profile of elution monitored at 280 nm.