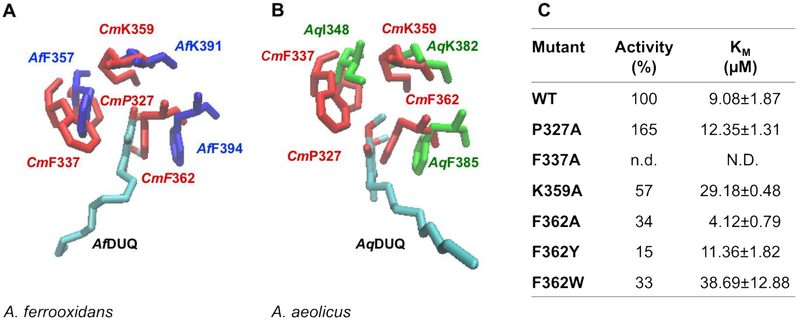
Figure 5. Residues involved in the putative quinone binding site.
C. maquilingensis SQR structural model (red) overlapped with (A) A. ferrooxidans SQR (Af, blue) and (B) A. aeolicus (Aq, green) SQR crystal structures, shows conserved residues surrounding the quinone substrate. (C) Activity and Km for DUQ of WT and mutants. 100% activity corresponds to 0.76 ± 0.03 μM of reduced DUQ per μM protein−1 per sec−1; n.d.: not detected; N.D.: not determined. Data are expressed as average ± SD of at least three independent experiments. Standard deviations for all the activity values were lower than 10 %.