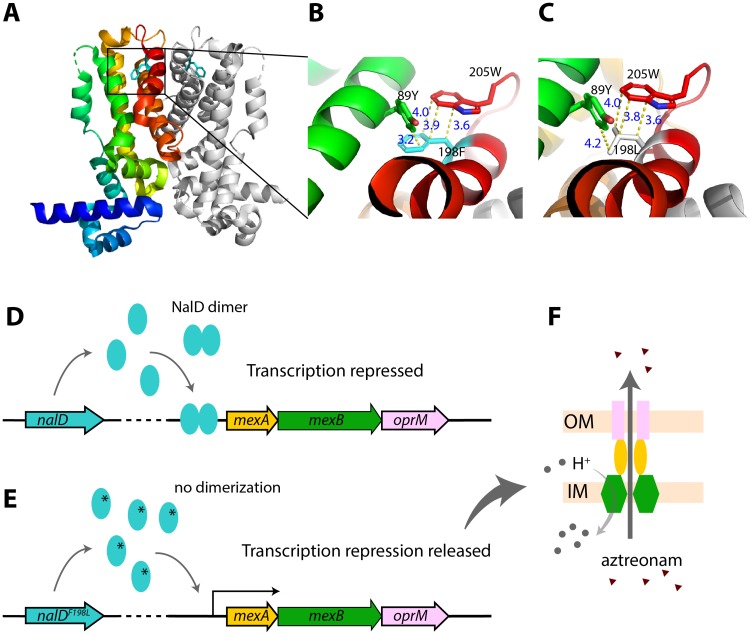
Fig 5. Molecular details of mechanism of acquired aztreonam resistance in the sepsis patient infected with P. aeruginosa.
(A) 3D structure of NalD dimer (PDB id: 5daj) with residue 198F (phenylalanine) shown as stick model. One copy of NalD is rainbow colored, while the other is in gray. The 10th helix is shown in red. (B) A closer look of residue 198F and possible interaction with two nearby aromatic residues, 89Y (tyrosin) and 205W (tryptophan). Those three residues are close together and could have aromatic interactions or a possible hydrogen bond (3.2 Å) between 198F and 89Y. 198F also aligns well with 205W, both of which have ring structure and could form a displaced pi stacking that stabilizes the NalD structure (3). (C) Prediction of mutation effect based on NalD structure. The mutation F198L would widen the distance between carbon groups and lose the pi-pi interaction, which could ultimately destabilize NalD dimerization. (D) Wild-type NalD dimer represses transcription of mexAB-oprM operon. (E) The mutation NalDF198L could interfere with dimerization, and de-repress transcription. (F) MexAB and OprM form an anti-porter system that exports aztreonam, increasing resistance [29].