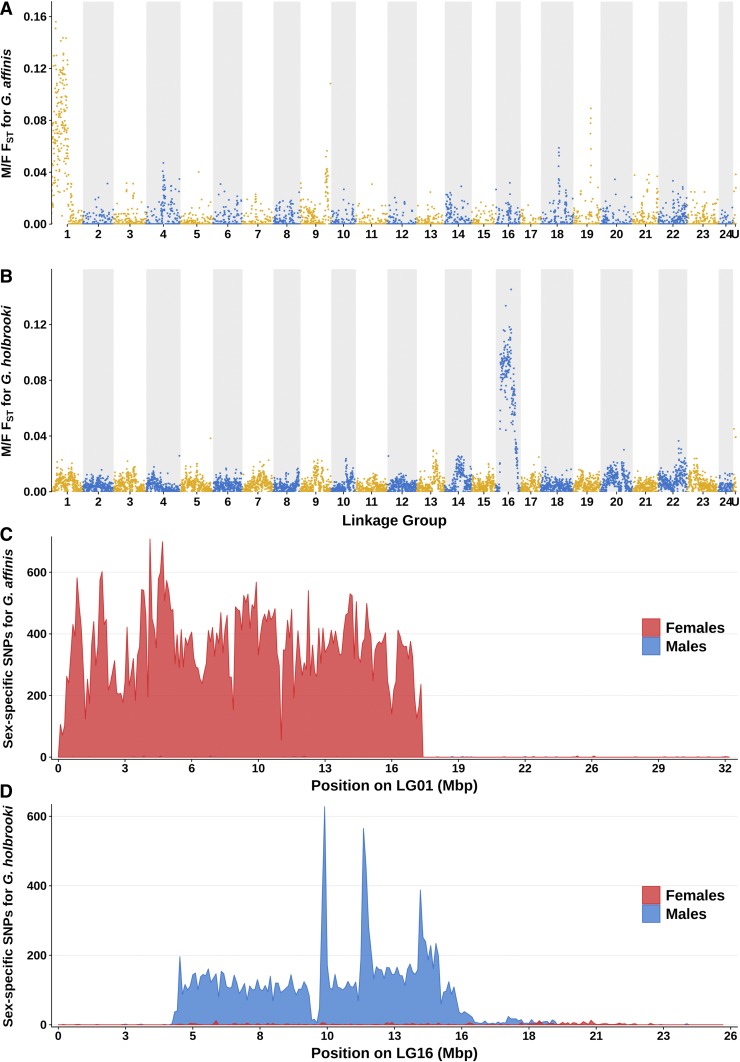
Figure 3.
Analysis of G. affinis and G. holbrooki pooled-sequencing reads aligned to the genome of X. maculatus. Male/female FST was computed in nonoverlapping 100 kb windows from G. affinis (A) and G. holbrooki (B) pooled-sequencing reads aligned to X. maculatus genome. Odd linkage groups (LGs) are colored in yellow with light background, and even LGs are colored blue with dark background. For G. affinis, a large region at the first half of LG1 shows a high male/female FST compared to the rest of the genome, while for G. holbrooki, a large region in the center of LG16 has the highest male/female FST. These results indicate that around half of LG1 of X. maculatus is homologous to the sex chromosome of G. affinis, while half of LG16 of X. maculatus is homologous to the sex chromosome of G. holbrooki. The number of female-specific SNPs (FSS, in red) and male-specific SNPs (MSS, in blue) was similarly computed, and results are shown for LG1 for G. affinis (C) and LG16 for G. holbrooki (D). For G. affinis, a region spanning from 0 to 17.7 Mb on LG1 was strongly and homogeneously enriched in FSS compared to the rest of the genome, while for G. holbrooki, the region enriched in MSS was located between 4.31 and 16.1 Mb on LG16, and showed three peaks of MSS density ∼10.2, 11.9, and 14 Mb.