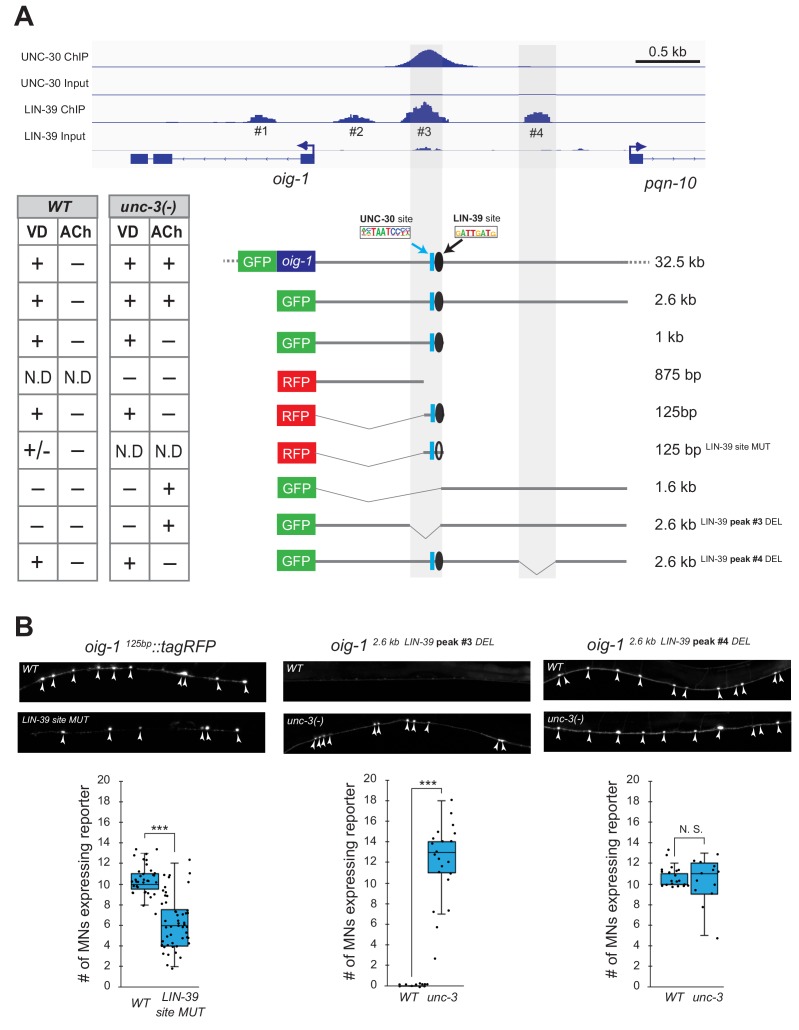
Figure 8. LIN-39 acts through distinct cis-regulatory elements to activate oig-1 expression in VD and VD-like neurons.
(A) ChIP-Seq tracks are shown for UNC-30 (the top two) and LIN-39 (the bottom two) on VD gene oig-1 locus. The UNC-30 data were obtained from Yu et al. (2017). The LIN-39 data come from the modENCODE project (Boyle et al., 2014). Four LIN-39 peaks are annotated with peak#3 largely overlapping with UNC-30 peak. The results of cis-regulatory analysis in both WT and unc-3 mutants are shown in the lower panel (aligned to the ChIP-seq tracks). Expression patterns of at least two transgenic lines were analyzed for each construct. ‘+' indicates consistent and bright expression in ventral nerve cord (VNC) MNs (either VD or cholinergic). ‘+/−' indicates consistent and bright expression in noticeable less number of VNC MNs. ‘−' indicates no or extremely dim expression in VNC MNs. ‘N.D.': Not determined. In the schematic of the transgenes, a known UNC-30 site is shown as a blue box and a bioinformatically predicted LIN-39 site is represented as a black circle (filled circle indicates the presence of the site while unfilled one indicates deletion of the site). MUT indicates deletion of the LIN-39 site and DEL indicates deletion of the respective LIN-39 peak region. (B) Images (top part) and quantifications (bottom part) of selected constructs in the cis-regulatory analysis shown in (A). Animals carrying the oig-1125bp::tagRFP (left panel) with the LIN-39 site deleted show reduced tagRFP reporter expression in VD neurons; animals carrying the oig-12.6kb LIN-39 peak #3 DEL (middle panel) ectopically express the reporter in cholinergic MNs of unc-3 mutants, but not in WT animals; animals carrying the oig-12.6kb LIN-39 peak #4 DEL (right panel) do not show ectopic reporter expression in unc-3-depleted MNs, but do show VD expression in both wild-type and unc-3 mutants. N > 12. ***p<0.001. N. S: not significant.