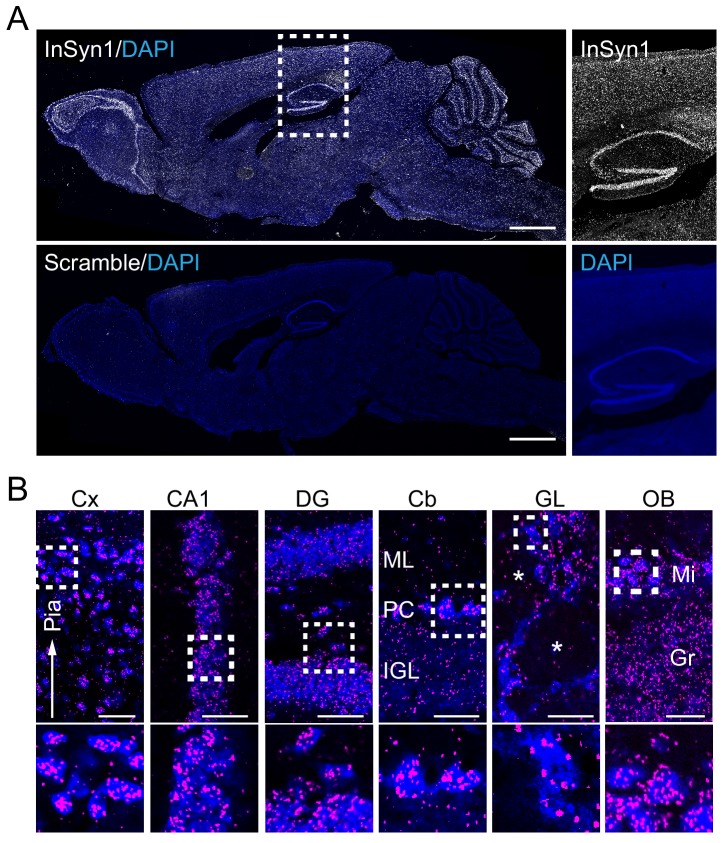
Figure 3. InSyn1 expression distribution in the mouse brain.
(A) InSyn1 mRNA (white) was detected throughout the adult mouse brain with strong signals in the hippocampus, cerebellum and olfactory bulb. Nissle stain (blue). Magnified images of the hippocampus and the cortex are shown. (B) Numerous clusters were found in cells in different layers of the cortex (Cx), pyramidal cell layers (CA1) and dentate gyrus granule cells (DG) in the hippocampus, Purkinje cells in the cerebellum (Cb), cells surrounding the glomerulus (GL) and in the mitral cell layer of the olfactory bulb (OB). Cx; cerebrum cortex, CA1; hippocampus CA1, DG; dentate gyrus, Cb; cerebellum, OB; olfactory bulb, GL; glomerular layer, Mi; mitral cell layer, Gr; granular cell layer, ML; molecular cell layer, PCL; Purkinje cell layer, IGL; internal granule layer. The asterisk represents the glomerulus. Scale bars, 1 mm (A), 50 um (B).