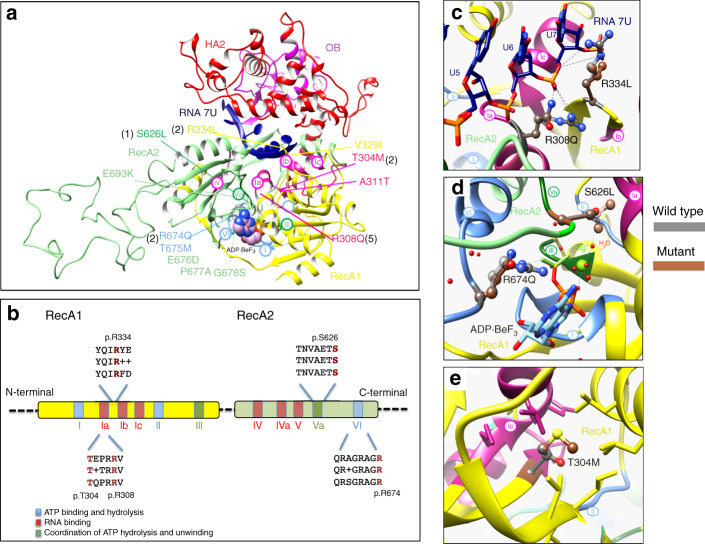
Fig. 2.
In silico modeling of DHX37 RecA1 and RecA2 pathogenic variants. (a) Functional domains of a homology model of DHX37 protein with U7-RNA (dark blue) and a nonhydrolyzable adenosine triphosphate (ATP) analog (ADP-BeF3, spheres). Domains are color-coded and labeled, conserved motifs are specified in circles, and the five disease-associated variants are indicated. The protein has four functional domains: two RecA-like domains, which are the helicase domains (RecA1: ATP-binding DEAH-box helicase, yellow, RecA2: C-terminal helicase, green); helicase associated 2 domain (HA2, red); and oligonucleotide/oligosaccharide-binding like domain (OB, pink). (b) Schematic diagram of the RecA-like domains in DEAH-box RNA helicases. Colors represent main helicase functions. Alignment of human (top) and Saccharomyces cerevisiae (bottom) showing the positions of recurrent variants causing 46,XY disorders/differences of sex development (DSD) with the consensus sequence shown in the middle. (c) Zoomed-in view of residue 308 and 334 with single-stranded RNA. (d) Residues 626 and 674 are shown to interact with the ATP analog. (e) Residue 304 is highlighted to be buried with a pocket within the RecA1 domain. Dashed lines within figure parts are shown for selected noncovalent polar interactions.