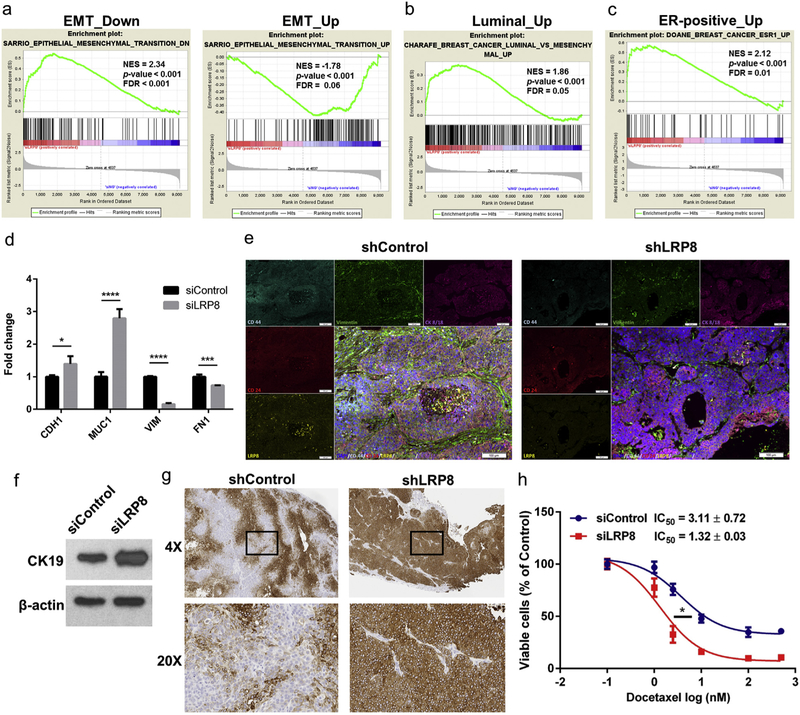
Fig. 6.
LRP8 knockdown shifts TNBC cells from a basal-mesenchymal state to a more differentiated luminal-epithelial state. a-c GSEA enrichment plots of EMT and luminal breast cancer gene sets in the LRP8 knockdown RNA-Seq data. a (left panel) Genes downregulated in mammary epithelial cells undergoing EMT (epithelial gene signature) were positively correlated with the LRP8 knockdown cells; (right panel) Genes upregulated in cells undergoing EMT (mesenchymal gene signature) were negatively correlated with the LRP8 knockdown cells. Genes upregulated in luminal breast cancer (b) and ER-positive breast cancer (c) were positively correlated with the LRP8 knockdown cells. NES: normalized enrichment score, FDR: false discovery rate. d qRT-PCR for the selected epithelial (CDH1: E-cadherin, MUC1: mucin 1) and mesenchymal (VIM: vimentin, FN1: fibronectin) genes in SUM149 with siLRP8 or siControl. Fold change between siLRP8 and siControl was calculated using the 2−ΔΔCt method and YWHAZ was used for normalization. The results were shown as mean ± S.D. (n = 3, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, Student’s t-Test). e Immunofluorescence staining of doxycycline-inducible control or LRP8 knockdown SUM149 xenograft tumors. f Western blot of CK19 in SUM149 cells with LRP8 knockdown by siRNA in vitro. g CK19 IHC in SUM149 tumors with control or LRP8 knockdown by doxycycline-inducible shRNA. h Relative cell survival of siControl or siLRP8 knockdown SUM149 cells treated with docetaxel (0, 1, 2.5, 10, 100, and 500 nM) for 72 h. The results were shown as mean ± S.D. (n = 3). IC50 of docetaxel was calculated and compared (*p < 0.05, Student’s t-Test).