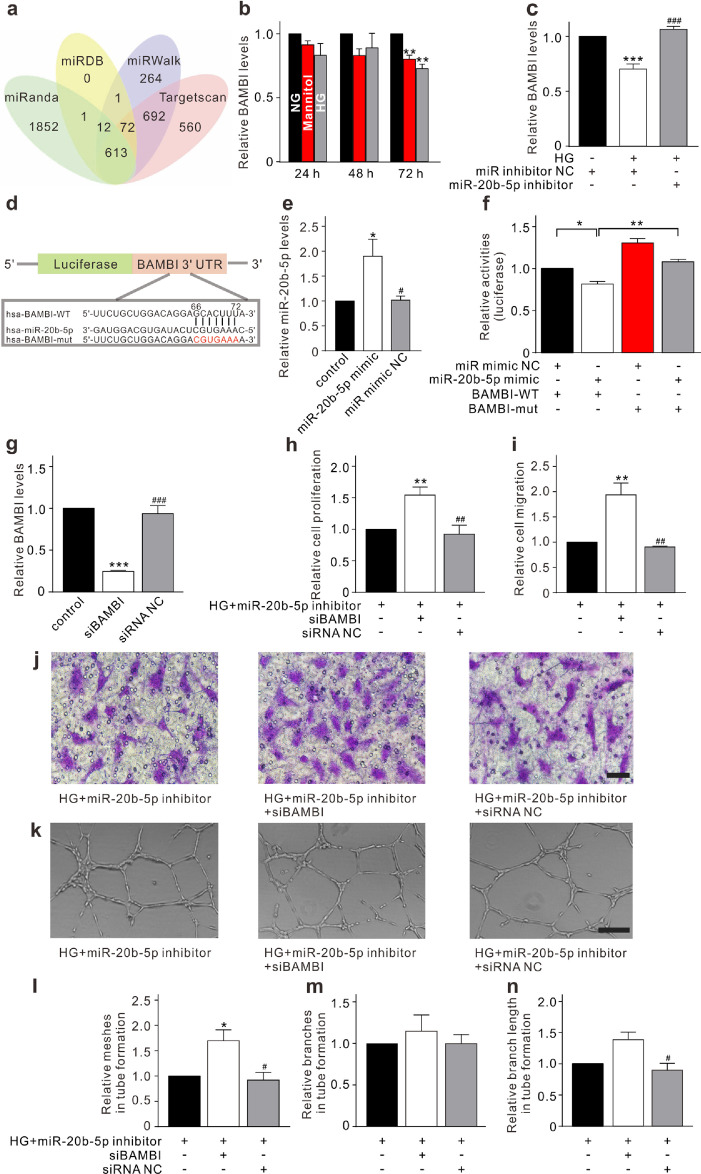
Fig. 2.
miR-20b-5p regulates HRMEC function through targeting BAMBI. (a) A Veen diagram illustrates the predicted target genes of miR-20b-5p from miRanda, miRDB, miRWalk, and Targetscan. (b) Changes of BAMBI levels, assayed by qRT-PCR, in HRMECs cultured in 5 mM glucose (normal glucose, NG), 5 mM glucose plus 25 mM mannitol (Mannitol), or 30 mM glucose (HG) medium for 24 h, 48 h, and 72 h, respectively. Data are normalized to corresponding NG groups. n = 3 for each group, ** p < 0.01 vs. NG. (c) Bar chart summarizing the changes of BAMBI levels in HRMECs transfected with miR inhibitor NC (control) or miR-20b-5p inhibitor (miR-20b-5p inhibitor group) with or without HG treatment. All data are normalized to control. n = 4 for each group, *** p < 0.001 vs. control, and ###p < 0.001 vs. HG. (d) Luciferase reporter vectors containing the wild-type or mutant 3′ UTR of the BAMBI were constructed according to the predicted binding sites of miR-20b-5p. (e) Bar chart summarizing the levels of miR-20b-5p in the 293T cells under different conditions. All data are normalized to control. n = 5 for each group, * p < 0.05 vs. control, and #p < 0.05 vs. miR-20b-5p mimic group. (f) Bar chart summarizing the changes of luciferase activities under different conditions. The HRMECs were co-transfected with luciferase reporter vector and miR-20b-5p mimic or miR mimic NC. n = 3 for each group, * p < 0.05, and ** p < 0.01. (g) Bar chart summarizing the changes of BAMBI levels in HRMECs transfected with siBAMBI or siRNA NC. All data are normalized to control. n = 3 for each group, *** p < 0.001 vs. control, and ###p < 0.001 vs. siBAMBI group. (h) Bar chart summarizing changes of HRMEC proliferation detected by CCK-8 assay. After transfected with miR-20b-5p inhibitor under HG conditions (control), the HRMECs were transfected with siBAMBI or siRNA NC. All data are normalized to control. n = 5 for each group, ** p < 0.01 vs. control, and ##p < 0.01 vs. siBAMBI group. (i, j) Bar chart summarizing the migration of HRMECs determined by Transwell assay under different conditions as shown in panel h (i). All data are normalized to control. n = 4 for each group, ** p < 0.01 vs. control, and ##p < 0.01 vs. siBAMBI group. Representative images are shown in panel j. Scale bar: 20 μm. (k–n) Representative images show the changes of capillary-like structure of HRMECs observed by Matrigel tube formation assay under different conditions as shown in panel h (k). Scale bar: 100 μm. Bar chart summarizing the changes of meshes (l), branches (m), and branching length (n). All data are normalized to control. n = 3 for each group; * p < 0.05 vs. control, and #p < 0.05 vs. siBAMBI group. All in vitro experiments: n = 3–5 biological replicates × 3 technical replicates. Data presented as means with error bars representing standard deviation (SD). Abbreviations: BAMBI = BMP and activin membrane bound inhibitor, CCK-8 = cell counting kit-8, HG = high glucose, HRMEC = human retinal microvascular endothelial cells, miR = microRNA, mut = mutant, NC = negative control, NG = normal glucose, qRT-PCR = quantitative reverse-transcription polymerase chain reaction, siBAMBI = small interfering BMP and activin membrane bound inhibitor, siRNA = small interfering RNA, UTR = untranslated region, WT = wild-type.