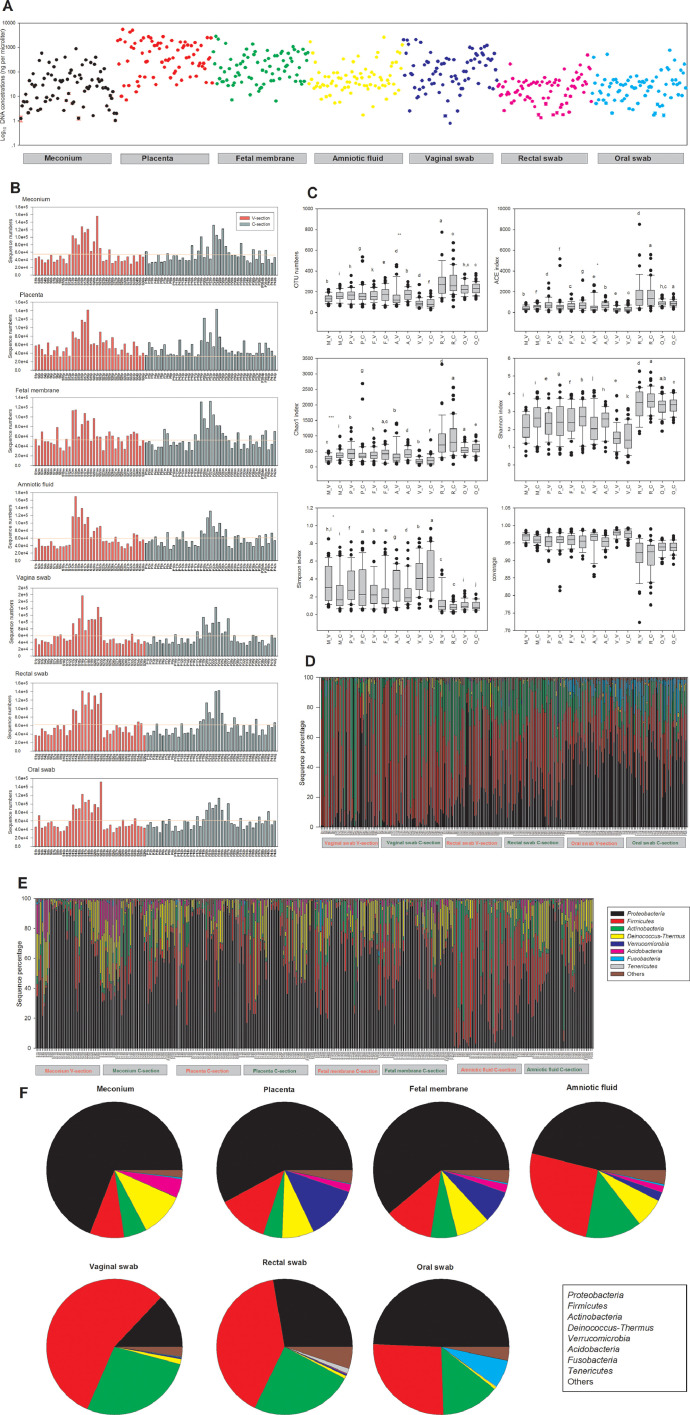
Fig. 1.
Microbiota abundance and composition in different mother-neonate pairs. (A) Concentration of total DNA extracted from each sample. (B) Number of sequences obtained by 16S rRNA gene sequencing. The numbers refer to individual mother-neonate pairs. The horizontal line reflects the average number of sequences for all samples. (C) Alpha-diversity box-whisker plots of OTU number, taxon richness (ACE and Chao1 indices) and diversity (Shannon–Wiener and Simpson indices) in samples analysed by 16S rRNA gene sequencing. The letters above the bars indicate the results of the Tukey HSD test following a significant 1-way ANOVA. Mean values that do not share the same letters were significantly different from each other (p < 0.05). (D) Relative abundances of bacterial taxa identified at the phylum level for each mother-related sample. (E) Relative abundances of bacterial taxa identified at the phylum level for each neonate-related sample. (F) Diversity of the most abundant bacterial taxa identified at the phylum level in different sample types (meconium, placenta, foetal membrane, amniotic fluid, vaginal swab, rectal swab and oral swab). Taxonomic assignment of 16S rRNA sequences was carried out with the SILVA SSU database v132 with a cutoff of 80% homology.