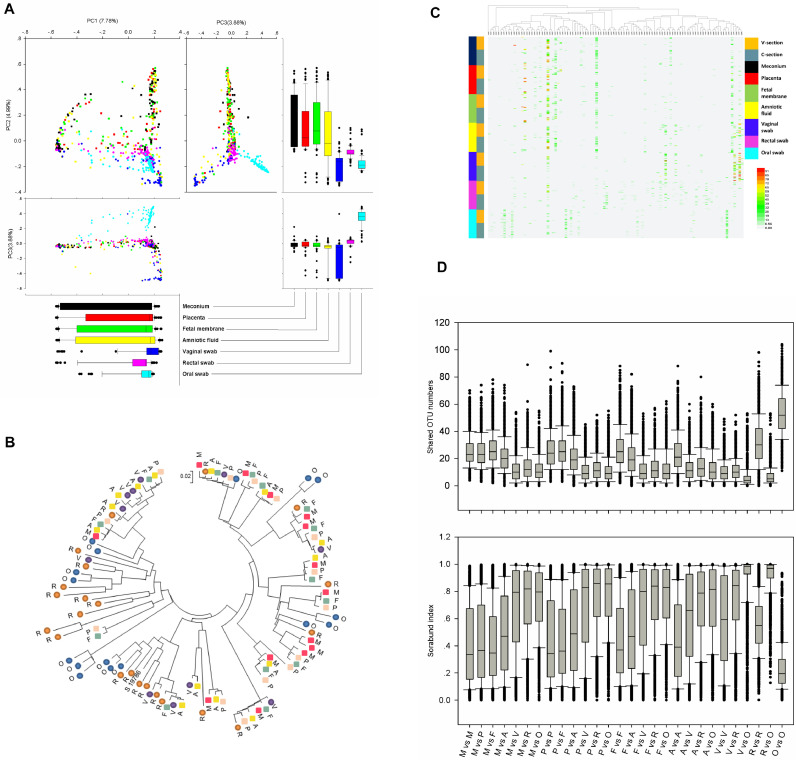
Fig. 2.
Comparison of the microbiomes of meconium, placenta, foetal membrane, amniotic fluid, vaginal swab, rectal swab and oral swab samples. (A) PCA plot based on the relative taxon abundance. Samples are marked by the group type. (B) Phylogenetic tree based on 16S rRNA gene sequences of major OTUs (more than 1% with a 0.03 cutoff) for each sample type. Meconium: red square, M; placenta: pink square, P; foetal membrane: grey-green square, F; amniotic fluid: yellow square, A; vaginal swab: purple circle, V; rectal swab: brown circle, R; oral swab: blue circle, O. (C) Heat map and phylogenetic analysis of the 100 most abundant OTUs (0.03 cutoff) in all samples. Vaginally delivered, yellow; C-section-delivered, grey blue; meconium, dark; placenta, red; foetal membrane, green; amniotic fluid, yellow; vaginal swab, blue; rectal swab, pink; oral swab, sky-blue. (D) Shared OTU numbers (0.03 cutoff) and sorabund index reflecting the similarities between different samples. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)