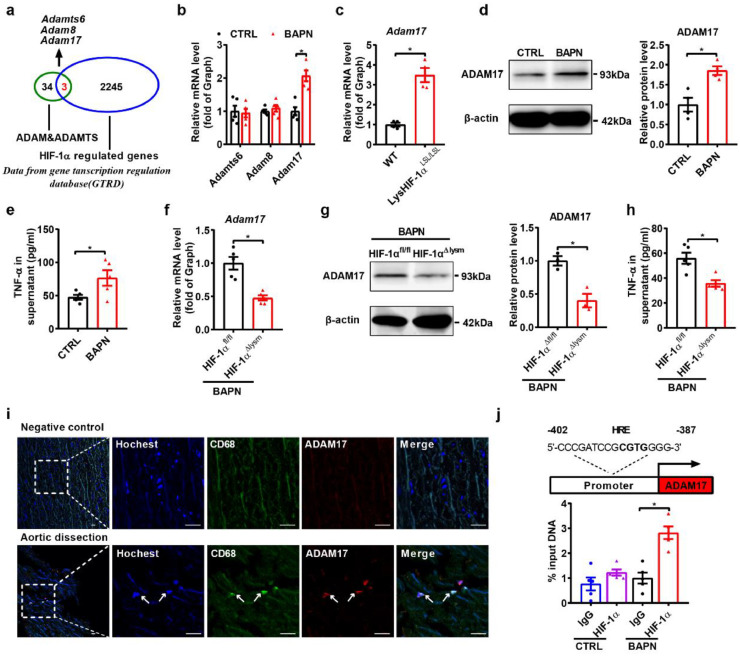
Fig. 4.
ADAM17 is identified as a target gene of HIF-1α in macrophages.
(a) Venn diagram illustrating overlap between HIF-1α-regulated genes and the genes of ADAM/ADAMTS, HIF-1α-regulated genes based on the Gene Transcription Regulation Database (GTRD).
(b) qPCR analysis of Adamts6, Adam8 and Adam17 mRNA levels in BAPN (1.2 mM, 12 h)-treated peritoneal macrophages (n = 5).
(c) qPCR analysis of the Adam17 mRNA level in peritoneal macrophages purified from wild-type and LysHIF-1αLSL/LSL mice (n = 4).
(d) Representative western blots and quantification for ADAM17 in BAPN-treated peritoneal macrophages (n = 3).
(e) ELISA analysis of the supernatant TNF-α level of BAPN-treated peritoneal macrophages (n = 4).
(f) qPCR analysis of the Adam17 mRNA level in peritoneal macrophages from HIF-1αfl/fl and HIF-1α△lysm mice treated with BAPN (n = 5).
(g) Representative western blots and quantification for ADAM17 in peritoneal macrophages (n = 3).
(h) ELISA analysis of the supernatant TNF-α level of peritoneal macrophages (n = 4).
(i) Immunofluorescence staining for macrophages (CD68, green) and ADAM17 (red) in transverse cryosections of aortas from patients with aortic dissection (n = 3). Nuclei were stained with Hoechst (blue). Scale bar, 25 µm.
(j) Schematic diagram of the HRE in the murine ADAM17 gene, and chromatin immunoprecipitation (ChIP) analysis of the occupancy of ADAM17 promoter regions by Hif1α (n = 5).
The data are presented as the mean ± SEM. Student's unpaired t-test was used for comparisons between two groups (b-h). Statistical significance is indicated by *P < 0.05, compared with the control group (b-e). *P < 0.05, compared with HIF-1αfl/fl mice (f-h); For ChIP analysis, the expression levels were normalized to the input levels; one-way ANOVA with Tukey's post hoc test (j) was performed. Statistical significance is indicated by *P < 0.05, compared with lgG (j).