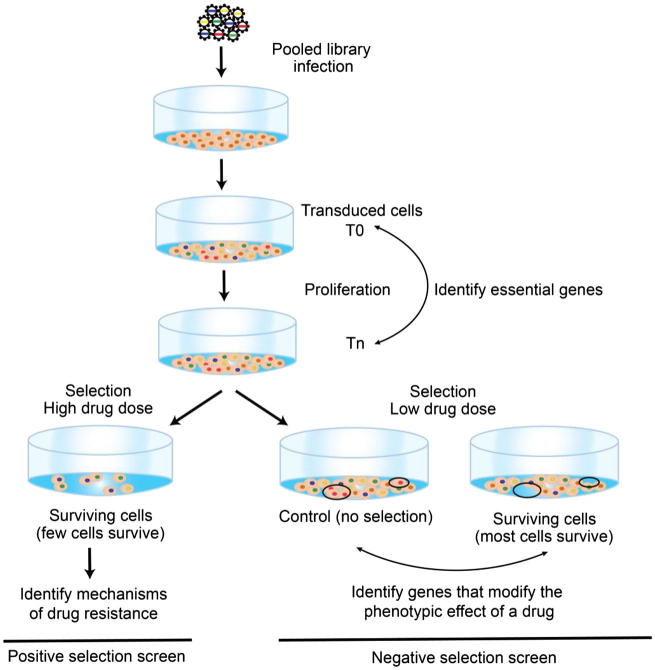
Fig. 2.
Experimental design for a whole genome CRISPR screen. In a pooled library CRISPR screen, cells are transduced with a pooled CRISPR library. Successfully transduced cells are sampled (T0) and grown for several doublings. At Tn cells are sampled again. Genomic DNA is extracted from T0 and Tn cells, PCR-amplified and sequenced using NGS. To identify essential genes (i.e. genes whose knock-out results in a fitness defect) abundance of each sgRNA at Tn is compared to abundance of each sgRNA at T0. Genome-wide CRISPR screens can be divided into two classes, positive and negative selection. In a positive screen, the goal is to identify those cells that survive post-selection (e.g. drug treatment). The selective pressure must be strong enough that most of the cells die, removing their sgRNAs from the population, and only a small fraction survives. After the surviving cells are collected, their plasmids are PCR-amplified and sequenced using NGS to identify their target gene. In a negative screen, the goal is to identify those cells that do not survive the selection mechanism. Two sets of cells are infected, one set is subject to selection (e.g. drug treatment) while the other set serves as a non-selected (i.e. non-treated) control. These two populations are then sequenced using NGS to determined which sgRNAs have been depleted by selection.