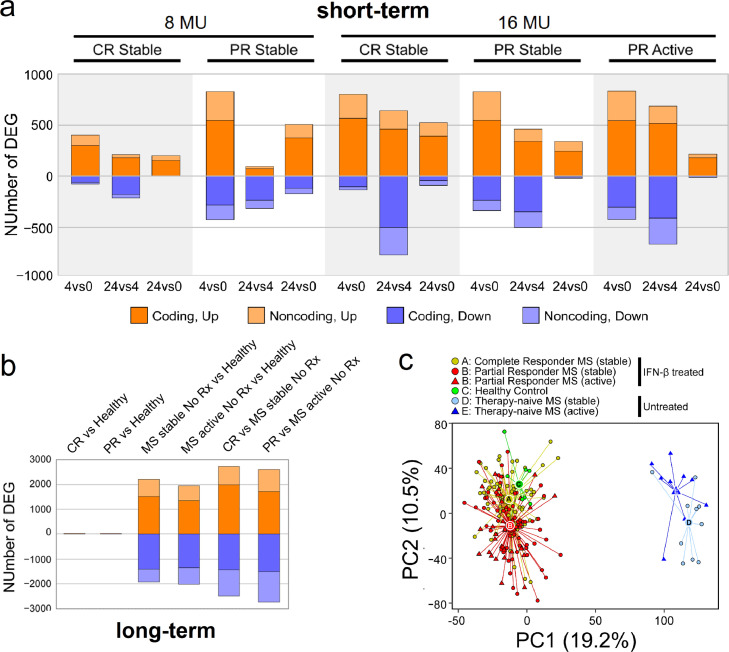
Fig. 1.
Differentially expressed genes (DEGs) after short-term and long-term IFN-β induction. (a) Short-term comparisons after 250 or 500 ug (8 or 16 MU) IFN-β injection. Panels include stable CR and stable and active PR without NAb to IFN-β. Comparisons on the x-axis include time 4 vs. 0 h, 24 vs. 0 h, and 24 vs. 4 h. The number of significant DEGs, with FDR-corrected p-value<0•05 and fold change ≥ 1•5 or ≤ −1•5, is shown on the y-axis. Genes upregulated are in orange; downregulated in blue. Coding DEG are in darker colour; non-coding genes in lighter colour. (b) Long-term compositions among baseline IFN-β-treated CR (stable) and PR (stable or active), therapy-naïve MS (stable or active), and healthy controls (HC), with FDR-corrected p-value<0•05, fold change ≥ 1•5 or ≤ −1•5. Colour coding same as in Fig. 1a. (c) Two-dimensional principal component analysis (PCA) of 227 samples using gene expression in all NAb-negative patients. Each circle or triangle represents one sample, for IFN-β-treated CR (A, yellow) and PR (B, red), 0, 4, and 24 h after injection, HC (C, green), therapy-naïve stable (D, light blue), and therapy-naïve active MS (E, dark blue). The variance of the five IFN-β-treated clinical groups is described by PC1 (19•2%) and PC2 (10•5%). The centroids of each group are labeled by letters A to E in larger circles or triangles. Euclidean distance between: CR and PR = 23•4; CR and HC = 15•4; PR and HC = 37•5; MS stable and MS active = 29•5; CR and MS stable = 132•5; PR and MS active = 124•7.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)