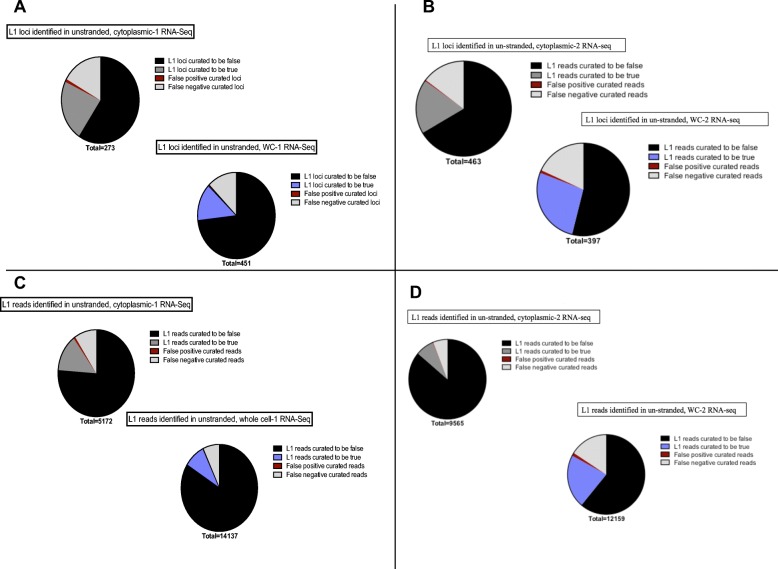
Fig. 5.
Curation required for data that is not strand specific. a-b Curation required by number of L1 loci in un-stranded cytoplasmic and whole cell RNA-seq data from replicates 1 and 2. Depicted are pie charts of the number of L1 loci that were curated to be truly or falsely expressed in non-strand-specific RNA-Seq data from whole cells or the cytoplasm. These curations were then compared to manual curation results of the matched strand-specific data in order to determine false positive and false negative calls. In black are the curated-to-be false loci, in light grey are the false negative calls determined when compared to strand-specific data, in red are the false positive calls made when compared to strand-specific data, in dark grey are the true loci identified in cytoplasmic RNA samples, and in purple are the true loci identified in whole-cell RNA samples. The number of total curated L1 loci is denoted beneath the pie charts. c-d Curation required by number of mapped reads to L1 loci in stranded cytoplasmic and whole cell RNA-seq data from replicates 1 and 2. Depicted are pie charts of the number of L1 mapped reads that were curated to be truly or falsely expressed in non-strand-specific RNA-seq data whole cells and cytoplasm. These curations were then compared to manual curation results of the matched strand-specific data in order to determine false positive and false negative calls. In black are the false reads, in light grey are the false negative calls determined when compared to strand-specific data, in red are the false positive calls made when compared to strand-specific data, in dark grey are the true L1 reads identified in cytoplasmic RNA samples, and in purple are the true L1 reads identified in whole cell RNA samples. The number of total curated L1 s reads is denoted beneath the pie charts