Figure 5.
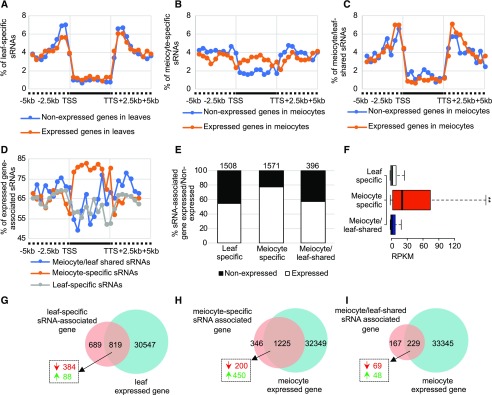
Soybean ms-sRNAs are positively correlated with meiotic gene expression. A, Distribution of ls-sRNAs around unexpressed and expressed genes in leaves. Intervals are partitioned into up- and down-stream regions (dashed line) and gene body (solid line) between TSSs and TTSs. B, Distribution of ms-sRNAs around unexpressed and expressed genes in meiocytes. C, Distribution of mls-sRNAs around unexpressed and expressed genes in meiocytes. D, Distribution of mls-, ms-, and ls-sRNAs around expressed genes. E, Histogram of mls-, ms-, and ls-sRNA–associated unexpressed and expressed genes. F, Gene expression value of mls-, ms-, and ls-sRNA–associated genes. **P value < 2.2e-16, Mann–Whitney test. G, Venn diagram showing 19% of differentially expressed genes (dashed box) are upregulated genes (green arrow, [q value < 0.05, log2 (fold change) > 1]) among ls-sRNA–associated genes, as opposed to downregulated (red arrow, [q value < 0.05, log2 (fold change) < −1]). H, Venn diagram showing 69% of the differentially expressed genes (dashed box) among the ms-sRNA–associated genes are upregulated (green arrow, [q value < 0.05, log2 (fold change) > 1]) as opposed to downregulated (red arrow, [q value < 0.05, log2 (fold change) < −1]). I, Venn diagram showing 41% of the differentially expressed genes (dashed box) among the mls-sRNA–associated genes are upregulated (green arrow, [q value < 0.05, log2 (fold change) > 1]) as opposed to downregulated (red arrow, [q value < 0.05, log2 (fold change) < −1]).