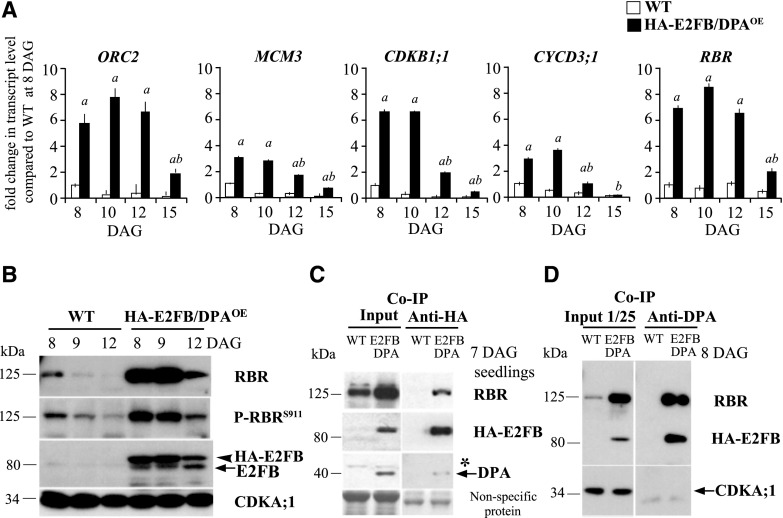
Figure 6.
Ectopic E2FB/DPA functions as transcriptional activator on cell cycle genes. A, The expression levels of ORC2, MCM3, CDKB1;1, CYCD3;1, and RBR were determined in wild-type (WT) and HA-E2FB/DPAOE seedlings by RT-qPCR. Developing first leaf pairs were analyzed at each time point, as indicated. Values represent the mean of fold change normalized to values of the relevant transcript from the wild type at 8 DAG, which was set arbitrarily at 1. Error bars indicate the sd; a, P < 0.05, statistical significance between the wild type and the transgenic line at a given time point; b, P < 0.05, significance between two consecutive time points determined using Student’s t test (n = 3, n > 100). Abbreviations of genes and the list of primers used in this study are listed in Supplemental Table S3. B, Protein level of RBR, P-RBRS911, HA-E2FB, and endogenous E2FB in the developing first leaf pairs of wild-type and HA-E2FB/DPAOE seedlings at 8, 9, and 12 DAG detected using anti-RBR, anti-P-RBRS911 (anti-P-Rb807/811), anti-E2FB, and anti-CDKA;1 antibodies in immunoblot assays. Note, the relative intensities of the RBR and P-RBRS911 protein bands are quantified in Supplemental Figure S6, F and G. C and D, Co-IP of HA-E2FB with RBR and DPA proteins in wild-type and HA-E2FB/DPAOE seedlings at 7 DAG (C) and in first leaf pairs at 8 DAG (D). Co-IP of RBR or HA-E2FB proteins with DPA was determined through immunoblot analysis with anti-RBR or anti-E2FB antibodies. One twenty-fifth of the IP from the extract was loaded as input. The asterisk indicates a nonspecific protein cross-reaction with the anti-DPA antibody in the input. In B and D, anti-CDKA;1 antibody was used as control. In C, nonspecific membrane-bound proteins stained by Coomassie-blue were used as loading control. The arrowhead in B indicates HA-E2FB and arrows mark the positions of endogenous E2FB, DPA, and CDKA;1 in B–D, respectively.