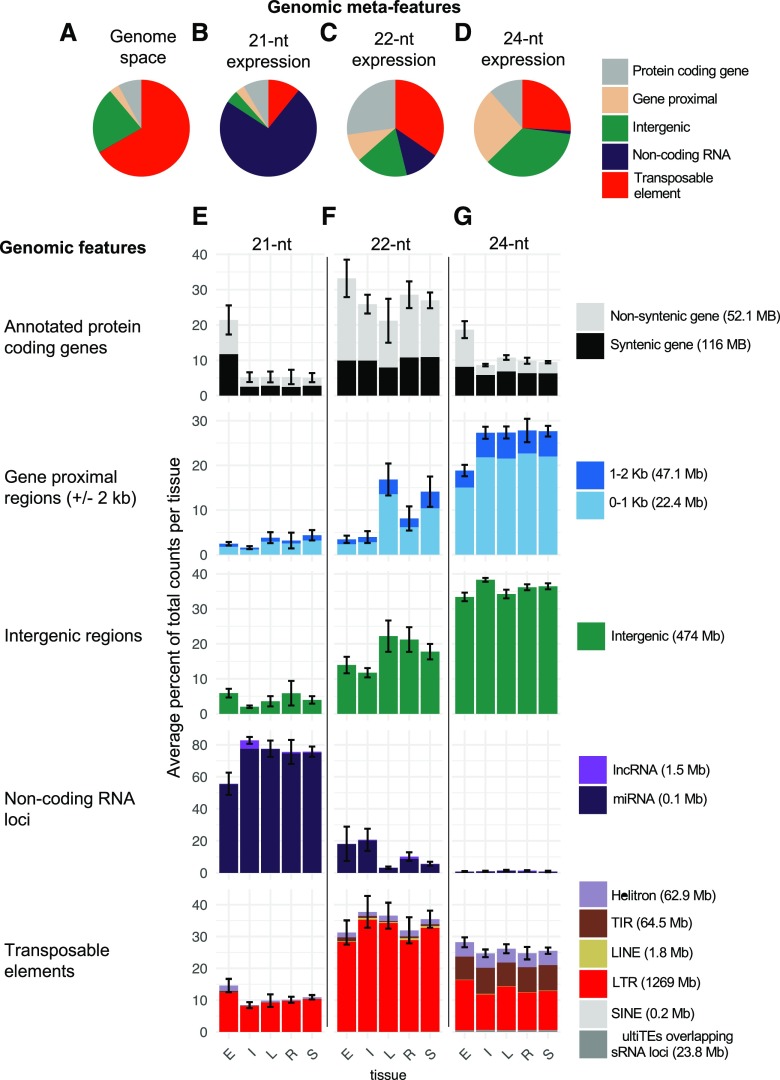
Figure 2.
Distribution of sRNA expression among the genomic features of the genome. A, Background genomic distribution of genomic features determined by annotating each 100-bp tile of the genome as noncoding RNA (e.g. miRNA and long noncoding RNA [lncRNA]), genic, gene-proximal (within 2 kb of a gene), TE, or intergenic using the genome reference B73 RefGen_v4 and annotation based on Gramene version 36 and miRbase release 22. B to D, Distribution of sRNA expression (CP5M) across meta-features for each sample averaged to determine the meta-feature distribution for each size class. E to G, Each meta-feature was then divided into constituent features, and the average distribution of sRNA expression was determined per tissue. In parentheses, the total Mb pairs of each genomic feature category in the genome are listed. The bars in the same vertical column add to 100%. Error bars denote sd among samples for the meta-feature category and represent the variation across the panel of inbred and hybrid genotypes. E = endosperm (n = 28), I = internode (n = 21), L = leaf (n = 18), R = seedling root (n = 20), and S = seedling shoot (n = 21). LINE, Long interspersed nuclear element; LTR, long terminal repeat; SINE, short interspersed nuclear element; TIR, terminal inverted repeat.