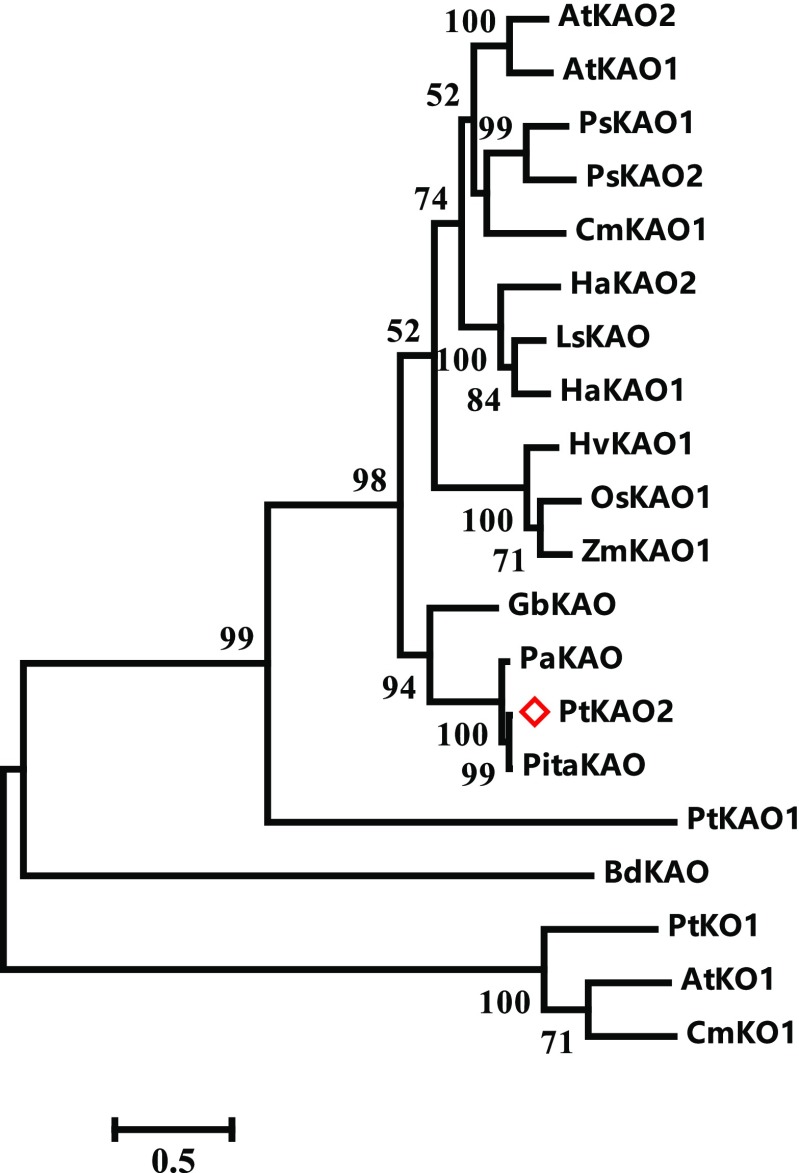
Figure 5.
Phylogenetic relationship of PtKAOs in P. tabuliformis and KAOs identified from angiosperms. PtKAO1, -2, and ZmKAO1 (Winkler and Helentjaris, 1995), OsKAO1 (Sakamoto et al., 2004), HvKAO1 (Helliwell et al., 2001), LsKAO (Sawada et al., 2008), HaKAO1 and 2 (Fambrini et al., 2011), PsKAO1 (Davidson et al., 2003), CmKAO1 (Helliwell et al., 2000), AtKAO1 and 2 (Helliwell et al., 2001), and the putative KAOs from other gymnosperms as Ginkgo biloba, Picea abies, and Pinus taeda were used to build the maximum likelihood tree. The bacteria BdKAO (Nett et al., 2017) and the related cytochrome P450 monooxygenase (CYP701A) ent-kaurene oxidases (KOs) were used as outgroup. The red diamond symbol indicates the P. tabuliformis gene that is identified in this study. The horizontal branch lengths are proportional to the estimated number of amino acid substitutions per residue. Bootstrap values were obtained from 1,000 bootstrap replicates.