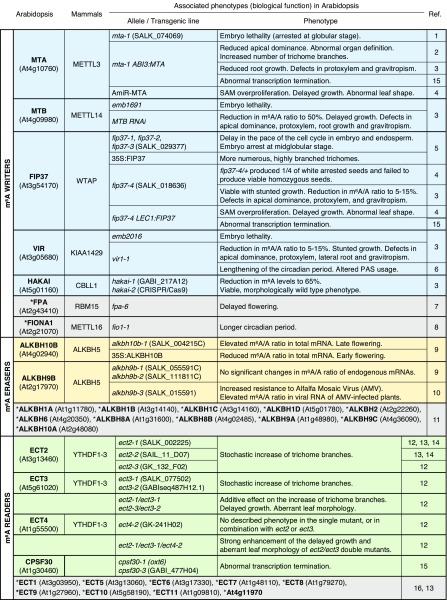
Figure 2.
Biological functions of the m6A pathway in plants as inferred by the mutant phenotype of its characterized components in Arabidopsis. The mammalian homologs of each factor is indicated. Arabidopsis bona fide writers are shaded in blue, erasers in orange, and readers in green. Arabidopsis genes marked with asterisks (as also labeled in Fig. 1) inside gray-shaded boxes are orthologs of m6A pathway components in other organisms or paralogs of such genes in Arabidopsis, but their possible or expected roles as m6A players in plants have not been verified experimentally. For this reason, columns 4 and 5 are omitted in the gray boxes corresponding to putative erasers and readers. Column 3 is also omitted for simplicity as it can be summarized as follows: AtALKBH proteins are homologs of the mammalian ALKBH1 to ALKBH8 and FTO family (Mielecki et al., 2012); all ECTs belong to the YTHDF clade (YTHDF1–YTHDF3 in mammals), while At4g11970 presents homology with the YTHDC clade (YTHDC1 and YTHDC2 in mammals; Scutenaire et al., 2018). In addition to knockout, knockdown, and overexpression lines, transgenic plants expressing point mutants of ECT2 (12–14), ECT3 (12), ECT4 (12), and CPSF30 (15) with impaired ability to bind m6A, or a catalytically inactive ALKBH10b (9), are also described in the indicated references and behave like null mutants for the phenotypes described in all cases. References are as follows: 1, Zhong et al., 2008; 2, Bodi et al., 2012; 3, Růžička et al., 2017; 4, Shen et al., 2016; 5, Vespa et al., 2004; 6, Parker et al., 2019; 7, Schomburg et al., 2001; 8, Kim et al., 2008; 9, Duan et al., 2017; 10, Martínez-Pérez et al., 2017; 11, Mielecki et al., 2012; 12, Arribas-Hernández et al., 2018; 13, Scutenaire et al., 2018; 14, Wei et al., 2018b; 15, Pontier et al., 2019; 16, Li et al., 2014a.