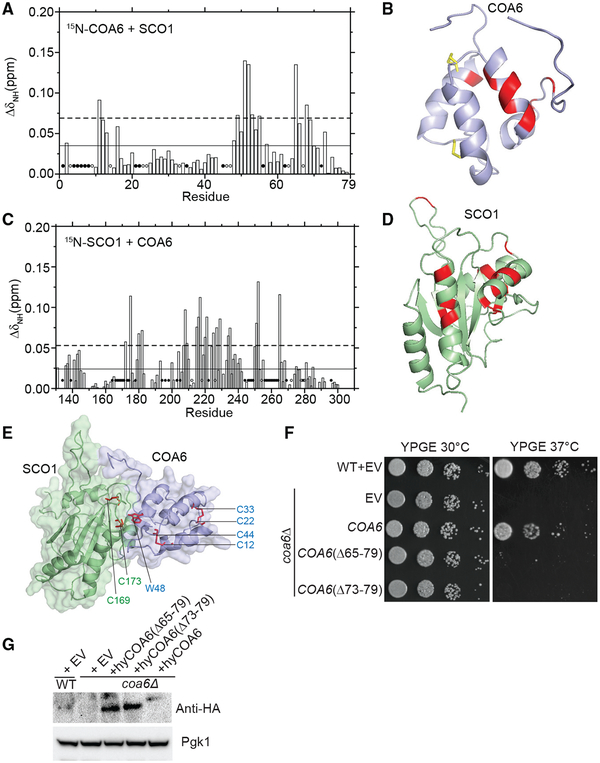
Figure 4. Mapping the Surfaces That Mediate Interactions between COA6 and SCO1.
(A) Average chemical shift perturbation of 15N-labeled COA6 upon addition of equimolar unlabeled SCO1. Horizontal solid line, mean chemical shift perturbation; dashed line, mean + SD of chemical shift perturbation for all residues; solid circles, unassigned residues; and open circles, residues with broadened peaks upon complex formation.
(B) COA6 residues with significant chemical shift perturbations are colored in red on a ribbon representation of COA6.
(C) Chemical shift perturbation of 15N-labeled SCO1 residues upon addition of equimolar unlabeled COA6, as displayed in (A).
(D) SCO1 residues with significant chemical shift perturbations are colored in red on a ribbon representation of SCO1.
(E) A HADDOCK docking model of the COA6:SCO1 complex generated using chemical shift perturbation (CSP) data. The side chain of the COA6 tryptophan (W48) and the indicated cysteine residues of COA6 and SCO1 are shown in stick representations in red.
(F) WT and coa6Δ cells transformed with empty vector (EV) or vector expressing either full-length or truncated hyCOA6. Transformants were serially diluted on agar plates and grown for 5 days.
(G) SDS-PAGE/western blot analysis of protein extracts from coa6Δ cells expressing wild-type or truncated mutants of hyCOA6. Pgk1 was used as loading control.