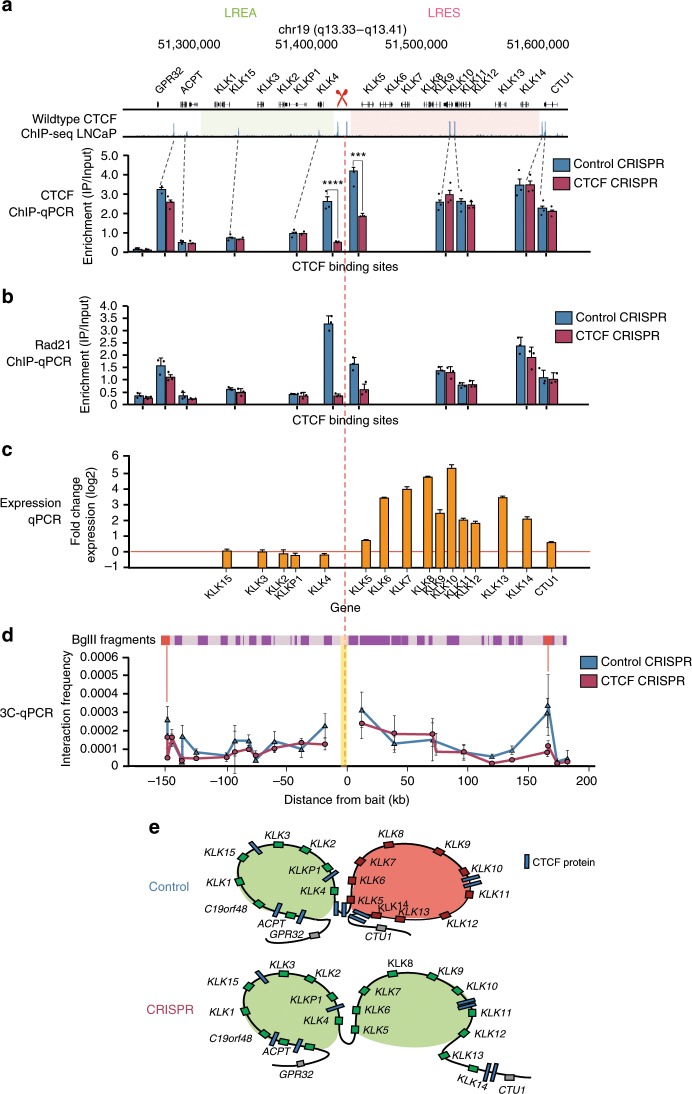
Fig. 5. CRISPR of persistent CTCF sites results in coordinate up-regulation of LRES genes.
a CTCF ChIP-qPCR assaying all CTCF-binding sites at KLK locus was performed on control and CRISPR cells and demonstrates reduction of CTCF binding at CRISPR sites but retained binding at all other CTCF sites in the region (* indicates where two-tailed t-test, p < 0.001; n = 2 biologically independent experiments). Source data are provided as a Source Data file. Error bars represent SE. Data for technical replicates n = 3 are overlaid as a dot blot. b RAD21 ChIP-qPCR performed after CRISPR of persistent sites shows that cohesin binding is maintained at persistent sites but depleted across the other CTCF sites in the region. Error bars represent SE. Data for technical replicates n = 3 are overlaid as a dot blot. Source data are provided as a Source Data file. c qPCR for KLK gene expression following CRISPR of two persistent sites at the KLK locus shows coordinate up regulation of previously silenced genes. Error bars represent SE. Source data are provided as a Source Data file. d Biological replicate #1 of 3C-qPCR at the KLK locus following CRISPR of persistent sites shows changes to looping intensity between the bait fragment (yellow bar) and its upstream and downstream interacting fragments (red lines). Bars above the graph illustrate BglII fragments across the region. Error bars represent SE between technical replicates ((x3); see Supplementary Fig. 6a). e Schematic representation of data in d demonstrates the weakening of interaction between the bait and the downstream anchor point and expression of the normally silenced genes in the LRES region following CRISPR of CTCF-persistent sites.