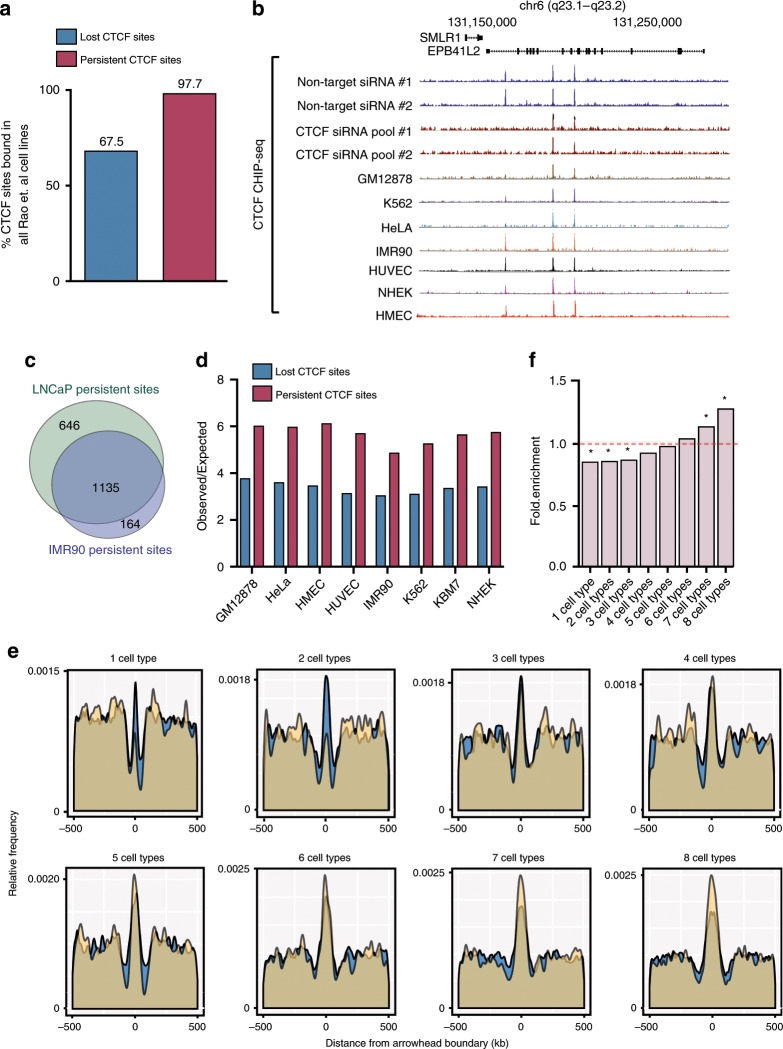
Fig. 7. Persistent sites are constitutively bound by CTCF in other cell types.
a Bar graph shows greater proportion of persistent sites are constitutively bound by CTCF in GM12878, K562, HeLA, IMR90, HUVEC, NHEK and HMEC cell lines when compared to lost sites. b CTCF ChIP-seq data for replicate LNCaP RNAi experiments and wildtype GM12878, K562, HeLA, IMR90, HUVEC, NHEK and HMEC cells shows persistent sites are constitutively bound by CTCF in other cell types at an example locus. c Venn diagram illustrating overlaps of LNCaP-persistent sites with IMR90-persistent sites. d Graph showing observed/expected ratio for lost (blue bars) and persistent sites (red bars) to overlap with domain boundaries in GM12878, K562, HeLA, IMR90, HUVEC, NHEK, HMEC and KBM7 cells. Comparing persistent to lost CTCF binding shows that persistent sites are more enriched to overlap domain boundaries. e Domain boundary data for GM12878, K562, HeLA, IMR90, HUVEC, NHEK, HMEC and KBM7 cells was subset based on whether each domain boundary was cell-type specific (present in 1 cell line), common (present in 2–7 cell lines) or cell-type constitutive (present in all 8 cell lines). Positional enrichment of lost and persistent CTCF sites was plotted for each subset. Lost sites are enriched at cell-type-specific domain boundaries. Persistent sites are enriched at cell-type constitutive domain boundaries (two-tailed t-test, *p = 0.02; n = 2 biologically independent experiments). f Enrichment of housekeeping genes at cell-type specific, cell-type common and cell-type constitutive TAD boundaries from Rao et al.4 (Hyper-geometric test, *p-value < 0.05 after Bonferroni correction).